Search Count: 4,919
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: RNA BINDING PROTEIN Ligands: ZN |
 |
Crystal Structure Of Glyoxylate/Hydroxypyruvate Reductase In Complex From Bacillus Subtilis With Formate At Near-Atomic Resolution
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2026-01-21 Classification: OXIDOREDUCTASE Ligands: MG, FMT, GOL |
 |
Crystal Structure Of Glyoxylate/Hydroxypyruvate Reductase In Complex From Bacillus Subtilis With Formate And Nadph At Near-Atomic Resolution
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2026-01-21 Classification: OXIDOREDUCTASE Ligands: MG, FMT, NAP, NA |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: MG, CL |
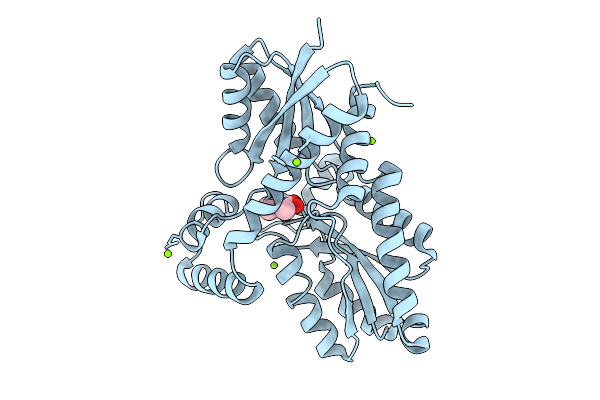 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: 9UM, MG, CL |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: 9UM, MG, MN, CL |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: MG, CL, 9UM, FE2 |
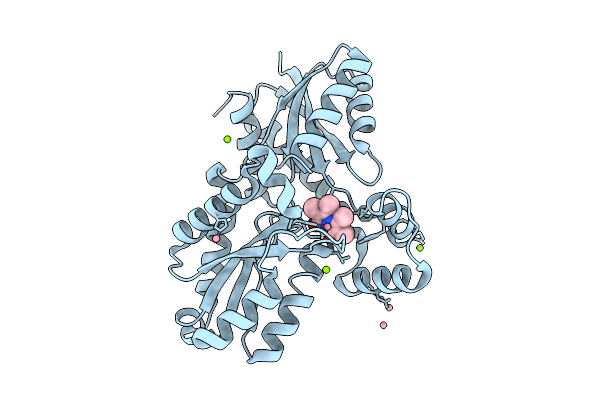 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: CO, MG, CL, 9UM |
 |
Ni(Ii)-Bound Bacillus Subtilis Cpfc (Hemh) Y13C Variant Modified With Bromobimane
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: 9UM, MG, NI, CL |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: 9UM, CU, MG, CL |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN, MG, 9UM |
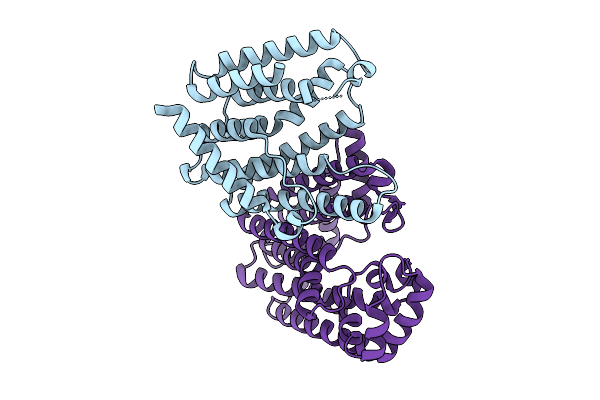 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2025-11-26 Classification: TRANSFERASE |
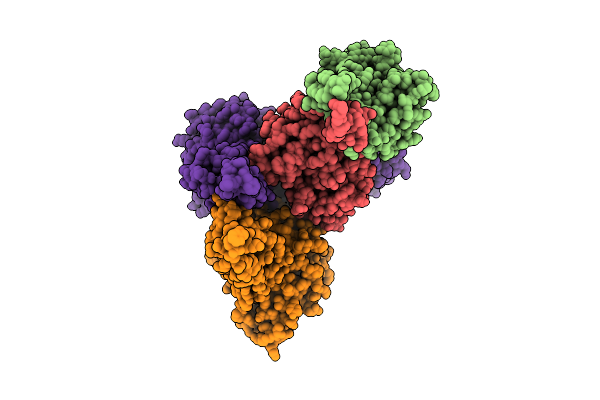 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: PROTEIN TRANSPORT |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Resolution:3.05 Å Release Date: 2025-11-12 Classification: STRUCTURAL PROTEIN |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Resolution:3.40 Å Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Resolution:2.50 Å Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: ADP, VO4, MG, UNL |
 |
Crystal Structure Of Bacillus Subtilis 168 L-Asparaginase Ii With Antileukemic Activity
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2025-10-08 Classification: ANTITUMOR PROTEIN |
 |
Organism: Salmonella enterica subsp. enterica serovar tennessee, Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: IMMUNE SYSTEM |
 |
Organism: Salmonella enterica subsp. enterica serovar tennessee, Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: IMMUNE SYSTEM |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Resolution:3.32 Å Release Date: 2025-09-24 Classification: STRUCTURAL PROTEIN |

