Search Count: 479
 |
Crystal Structure Of A Snoal-Like Domain-Containing Protein From Mycobacterium Ulcerans (Orthorhombic C Form)
Organism: Mycobacterium ulcerans agy99
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: ISOMERASE Ligands: PG4 |
 |
Crystal Structure Of A Snoal-Like Domain-Containing Protein From Mycobacterium Ulcerans
Organism: Mycobacterium ulcerans agy99
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: ISOMERASE Ligands: PO4, NA |
 |
Laser Excitation Effects On Br: Reprocessed Dark Dataset Recorded From Nango Et Al.
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Laser Excitation Effects On Br: Reprocessed Extrapolated 16 Ns Light Dataset Recorded At 0.04 Gw/Cm2 From Nango Et Al.
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Laser Excitation Effects On Br: Extrapolated 6 Ps Light Dataset Recorded At 342 Gw/Cm2 At Swissfel
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Laser Excitation Effects On Br: Extrapolated 6 Ps Light Dataset Recorded At 2493 Gw/Cm2 At Swissfel
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Laser Excitation Effects On Br: Reprocessed Extrapolated 10Ps Light Dataset Recorded At 525 Gw/Cm2 From Nogly Et Al.
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Laser Excitation Effects On Br: Extrapolated 10Ps Light Dataset Recorded At 1281 Gw/Cm2 At Sacla (+100Ps, 1Ns, 10Ns)
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
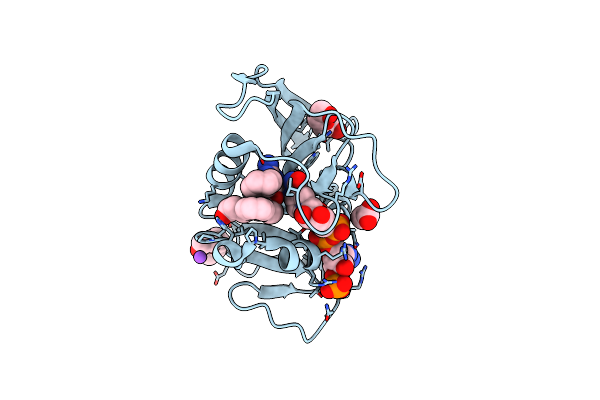
Crystal Structure Of Dihydrofolate Reductase (Dhfr) From Mycobacterium Ulcerans Agy99 In Complex With Nadp And Inhibitor Mam881
Organism: Mycobacterium ulcerans
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-08-28 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: DQI, NAP |
 |
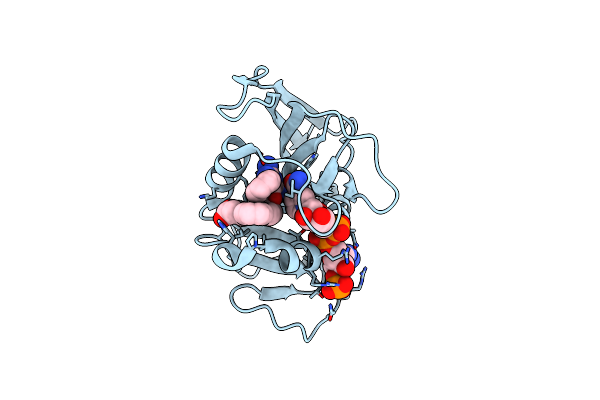
Crystal Structure Of Dihydrofolate Reductase (Dhfr) From Mycobacterium Ulcerans Agy99 In Complex With Nadp And Inhibitor Mam907
Organism: Mycobacterium ulcerans
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-08-28 Classification: OXIDOREDUCTASE/INHIBITOR |
 |
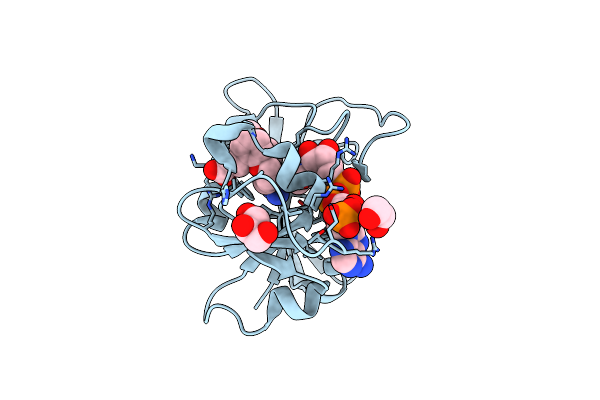
Crystal Structure Of Dihydrofolate Reductase (Dhfr) From Mycobacterium Ulcerans Agy99 In Complex With Nadp And Inhibitor Mam758
Organism: Mycobacterium ulcerans
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-08-28 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: NAP, HIJ |
 |
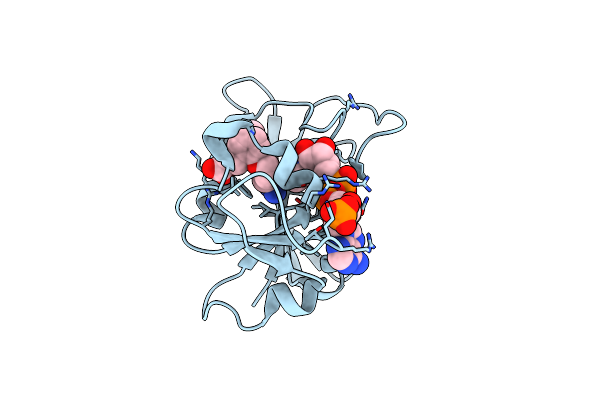
Crystal Structure Of Dihydrofolate Reductase (Dhfr) From Mycobacterium Ulcerans Agy99 In Complex With Nadp And Inhibitor Mam738
Organism: Mycobacterium ulcerans
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2023-12-13 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: NAP, XJB, GOL, EDO |
 |
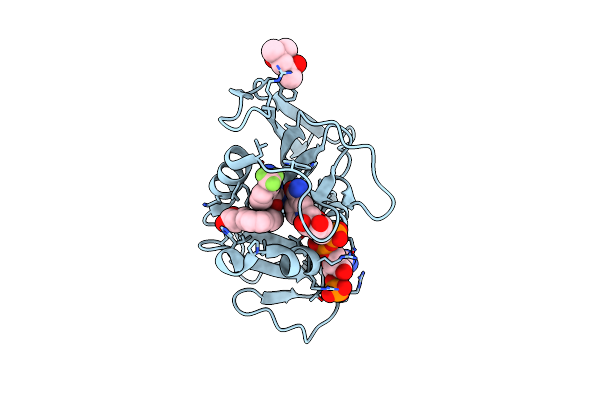
Crystal Structure Of Dihydrofolate Reductase (Dhfr) From Mycobacterium Ulcerans Agy99 In Complex With Nadp And Inhibitor Mam777
Organism: Mycobacterium ulcerans
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2023-12-13 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: NAP, XJN |
 |
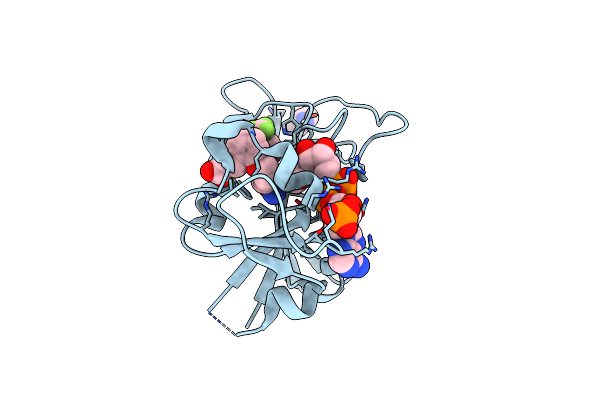
Crystal Structure Of Dihydrofolate Reductase (Dhfr) From Mycobacterium Ulcerans Agy99 In Complex With Nadp And Inhibitor Mam845
Organism: Mycobacterium ulcerans
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2023-12-13 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: XJT, MPD, NAP, BR |
 |
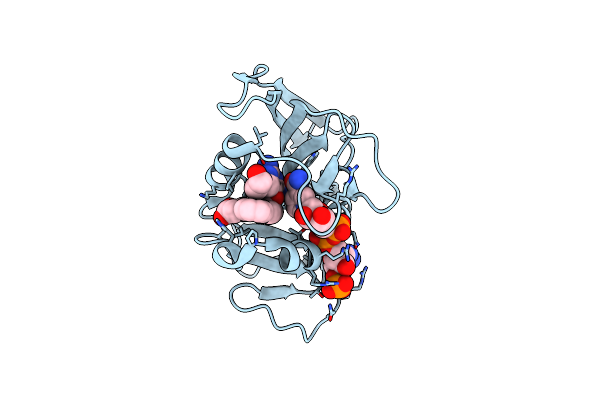
Crystal Structure Of Dihydrofolate Reductase (Dhfr) From Mycobacterium Ulcerans Agy99 In Complex With Nadp And Inhibitor Mam846
Organism: Mycobacterium ulcerans
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2023-12-13 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: NAP, XJZ, IMD |
 |
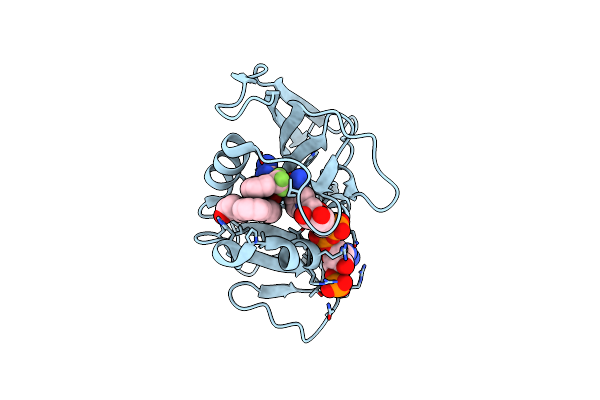
Crystal Structure Of Dihydrofolate Reductase (Dhfr) From Mycobacterium Ulcerans Agy99 In Complex With Nadp And Inhibitor Mam787
Organism: Mycobacterium ulcerans
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2023-12-13 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: NAP, XK8 |
 |
Crystal Structure Of Dihydrofolate Reductase (Dhfr) From Mycobacterium Ulcerans Agy99 In Complex With Nadp And Inhibitor Mam851
Organism: Mycobacterium ulcerans
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2023-12-13 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: NAP, XKI |

