Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
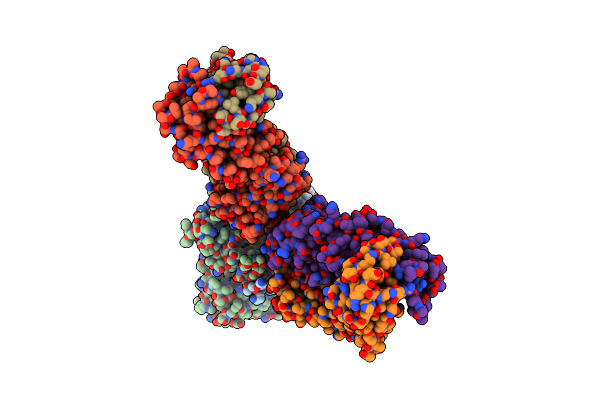 |
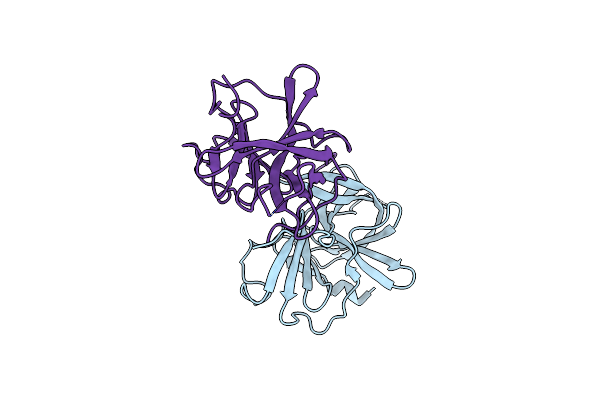
Mouse Norovirus Protruding Domain Complexed With Neutralizing Fab Fragment From Mab A6.2
Organism: Murine norovirus 1, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2021-04-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
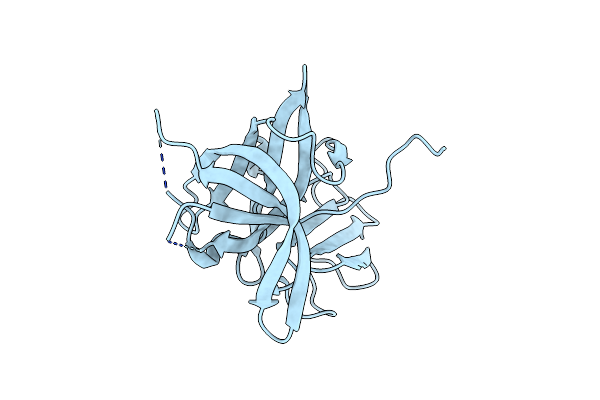
Crystal Structure Of Murine Norovirus P Domain In Complex With Nanobody Nb-5867
Organism: Murine norovirus 1, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2020-04-22 Classification: VIRAL PROTEIN Ligands: EDO |
 |
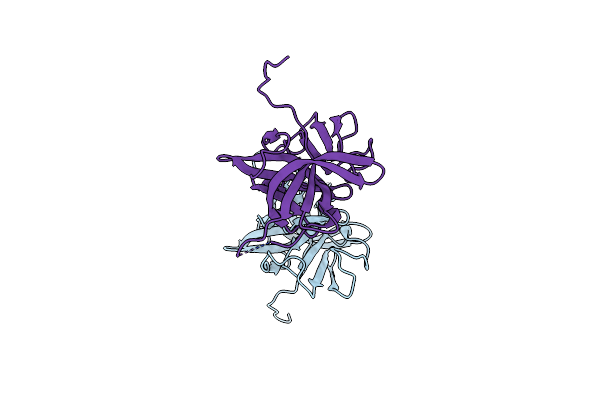
Crystal Structure Of Murine Norovirus P Domain In Complex With Nanobody Nb-5820
Organism: Murine norovirus 1, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2020-04-22 Classification: VIRAL PROTEIN Ligands: EDO, TRS |
 |
Crystal Structure Of Murine Norovirus P Domain In Complex With Nanobody Nb-5853
Organism: Murine norovirus 1, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2020-04-22 Classification: VIRAL PROTEIN Ligands: MG, EDO |
 |
Crystal Structure Of Murine Norovirus P Domain In Complex With Nanobody Nb-5829 And Glycochenodeoxycholate (Gcdca)
Organism: Murine norovirus 1, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-04-22 Classification: VIRAL PROTEIN Ligands: EDO, CHO, MG |
 |
Murine Norovirus Protruding Domain (Cw3 Strain) In Complex With The Cd300Lf Receptor And Glycochenodeoxycholate (Gcdca)
Organism: Murine norovirus 1, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-08-14 Classification: VIRAL PROTEIN Ligands: MG, CHO, NA, EDO |
 |
Bile Salts Alter The Mouse Norovirus Capsid Conformation; Possible Implications For Cell Attachment And Immune Evasion.
Organism: Murine norovirus 1
Method: ELECTRON MICROSCOPY Release Date: 2019-08-07 Classification: VIRUS |
 |
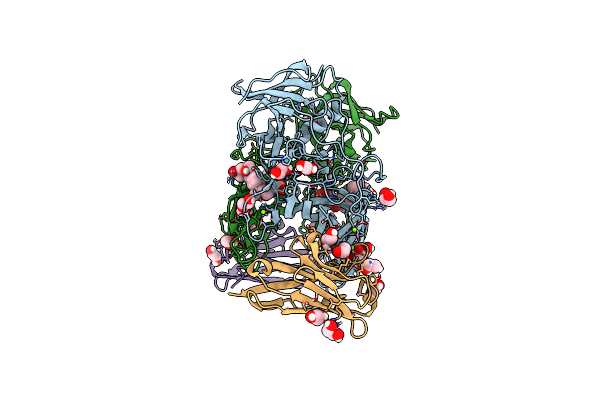
Crystal Structure Of The Murine Norovirus Vp1 P Domain In Complex With The Cd300Lf Receptor
Organism: Murine norovirus 1, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-09-12 Classification: VIRAL PROTEIN/LIPID BINDING PROTEIN Ligands: EDO, CL, MG |
 |
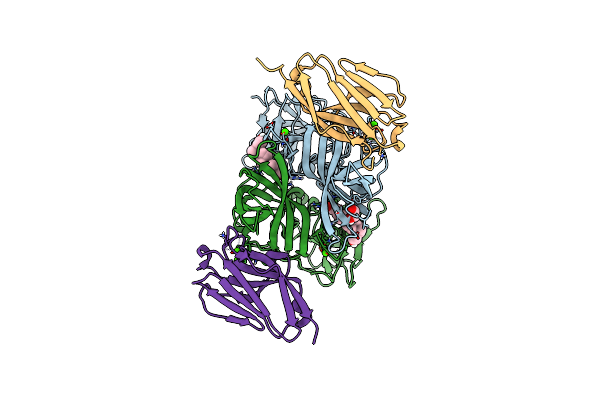
Crystal Structure Of The Murine Norovirus Vp1 P Domain In Complex With The Cd300Lf Receptor And Glycochenodeoxycholic Acid
Organism: Murine norovirus 1, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-09-12 Classification: VIRAL PROTEIN Ligands: MG, EDO, CHO |
 |
Crystal Structure Of The Murine Norovirus Vp1 P Domain In Complex With The Cd300Lf Receptor And Lithocholic Acid
Organism: Murine norovirus 1, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-09-12 Classification: VIRAL PROTEIN Ligands: 4OA, CA, EDO |
 |
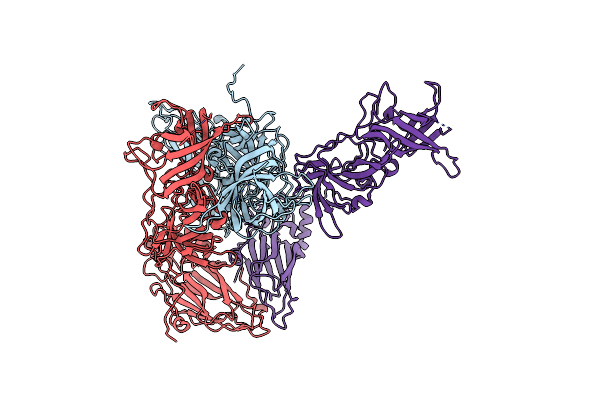
Structural Insight Into The Interaction Between Rna Polymerase And Vpg For Norovirus Replication
Organism: Murine norovirus 1, Murine norovirus
Method: X-RAY DIFFRACTION Resolution:3.14 Å Release Date: 2018-07-18 Classification: VIRAL PROTEIN |
 |
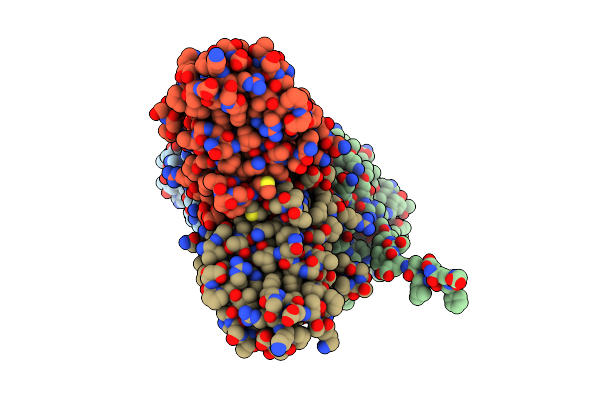
Mouse Norovirus Model Using The Crystal Structure Of Mnv P Domain And The Norwalkvirus Shell Domain
Organism: Norwalk virus, Murine norovirus 1
Method: ELECTRON MICROSCOPY Release Date: 2018-04-04 Classification: VIRUS |
 |
Crystal Structure Of The Murine Norovirus Ns6 Protease (Inactive C139A Mutant) With A C-Terminal Extension To Include Residue P1 Prime Of Ns7
Organism: Murine norovirus 1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-02-18 Classification: HYDROLASE Ligands: IMD |
 |
Crystal Structure Of The Murine Norovirus Ns6 Protease (Inactive C139A Mutant) With A C-Terminal Extension To Include Residues P1 Prime - P2 Prime Of Ns7
Organism: Murine norovirus 1
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-02-18 Classification: HYDROLASE |
 |
Crystal Structure Of The Murine Norovirus Ns6 Protease (Inactive C139A Mutant) With A C-Terminal Extension To Include Residues P1 Prime - P4 Prime Of Ns7
Organism: Murine norovirus 1
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2015-02-18 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of A Chimeric Murine Norovirus Ns6 Protease (Inactive C139A Mutant) In Which The P4-P4 Prime Residues Of The Cleavage Junction In The Extended C-Terminus Have Been Replaced By The Corresponding Residues From The Ns2-3 Junction.
Organism: Murine norovirus 1
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2015-02-18 Classification: VIRAL PROTEIN |
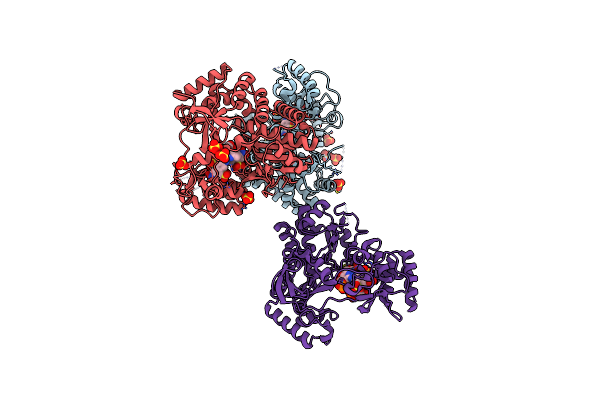 |
Organism: Murine norovirus 1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-11-05 Classification: VIRAL PROTEIN/REPLICATION INHIBITOR Ligands: 20V, SO4 |
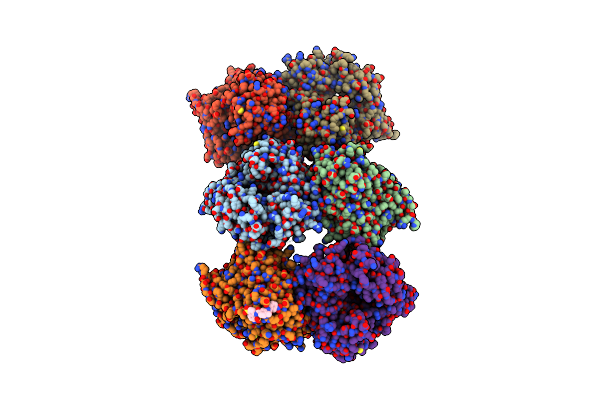 |
Murine Norovirus Rna-Dependent-Rna-Polymerase In Complex With Compound 6, A Suramin Derivative
Organism: Murine norovirus 1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-10-15 Classification: VIRAL PROTEIN/TRANSCRIPTION INHIBITOR Ligands: 2NG, MG |
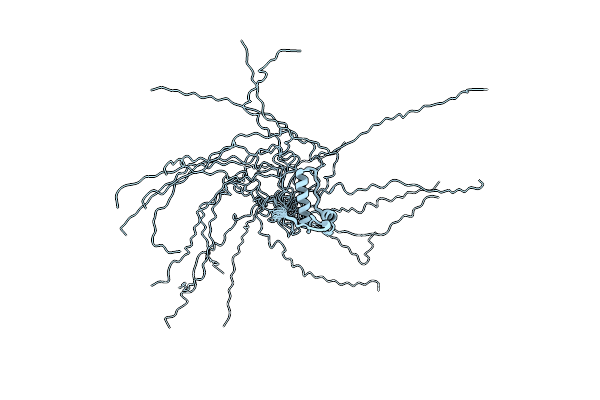 |
Backbone 1H, 13C, And 15N Chemical Shift Assignments For Murine Norovirus Ns1/2 D94E Mutant
Organism: Murine norovirus 1
Method: SOLUTION NMR Release Date: 2013-12-18 Classification: HYDROLASE |
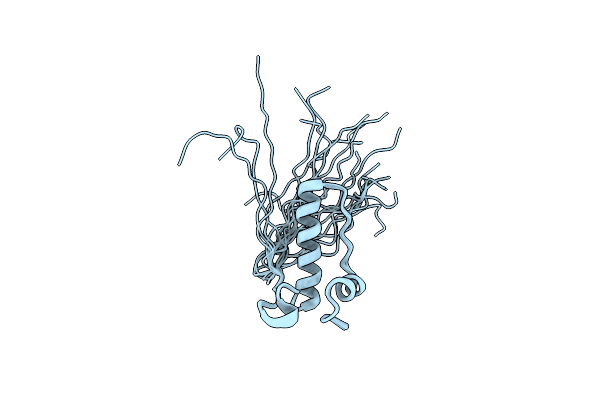 |
Backbone 1H, 13C, And 15N Chemical Shift Assignments For Murine Norovirus Ns1/2 Cw3 Wt
Organism: Murine norovirus 1
Method: SOLUTION NMR Release Date: 2013-12-18 Classification: HYDROLASE |