Search Count: 272
 |
Connectase T1A C192S Mutant From Methanocaldococcus Mazei With Peptide Substrate
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2025-01-15 Classification: LIGASE |
 |
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2025-01-01 Classification: LIGASE Ligands: 1PE, NA, GOL, EDO |
 |
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2024-12-25 Classification: LIGASE |
 |
Organism: Methanosarcina mazei
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: RNA |
 |
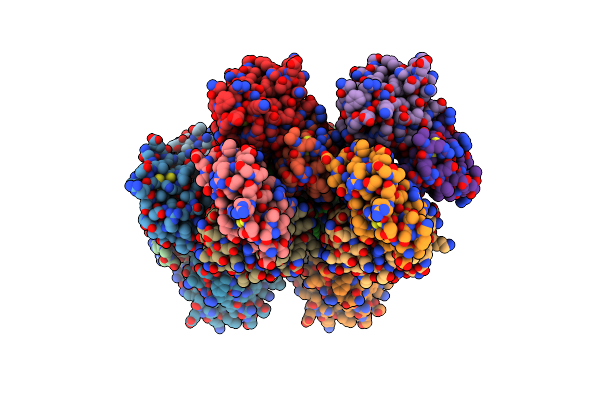
Cryo-Em Structure Of The Methanosarcina Mazei Apo Glutamin Synthetase Structure: Dodecameric Form
Organism: Methanosarcina mazei
Method: ELECTRON MICROSCOPY Release Date: 2023-11-15 Classification: LIGASE Ligands: MG |
 |
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-11-01 Classification: LIGASE Ligands: ANP, 6CV, MG |
 |
Pylrs C-Terminus Domain Mutant Bound With L-3-Trifluoromethylphenylalanine And Ampnp
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-11-01 Classification: LIGASE Ligands: MG, ANP, FX9 |
 |
Pylrs C-Terminus Domain Mutant Bound With D-3-Trifluoromethylphenylalanine And Ampnp
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-11-01 Classification: LIGASE Ligands: ANP, MG, FXC |
 |
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-11-01 Classification: LIGASE Ligands: ANP, MG, FXF |
 |
Pylrs C-Terminus Domain Mutant Bound With D-3-Chlorophenylalanine And Ampnp
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-11-01 Classification: LIGASE Ligands: MG, ANP, FXL |
 |
Pylrs C-Terminus Domain Mutant Bound With L-3-Chlorophenylalanine And Ampnp
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-11-01 Classification: LIGASE Ligands: ANP, MG, FCL |
 |
Organism: Arabidopsis thaliana, Aster yellows witches'-broom phytoplasma (strain aywb)
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2023-07-19 Classification: PLANT PROTEIN |
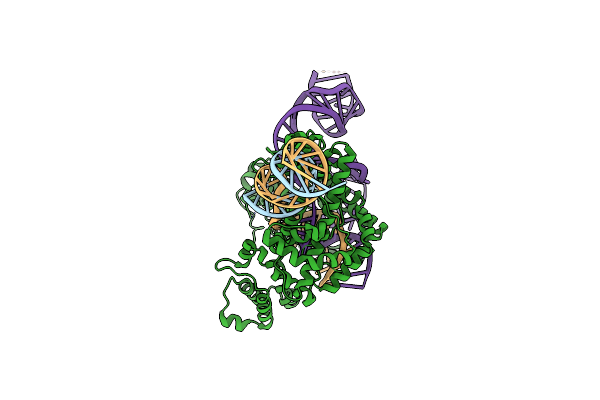 |
Structure Of The Spizellomyces Punctatus Fanzor (Spufz) In Complex With Omega Rna And Target Dna
Organism: Methanosarcina mazei, Spizellomyces punctatus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: RNA BINDING PROTEIN/RNA/DNA Ligands: MG, ZN |
 |
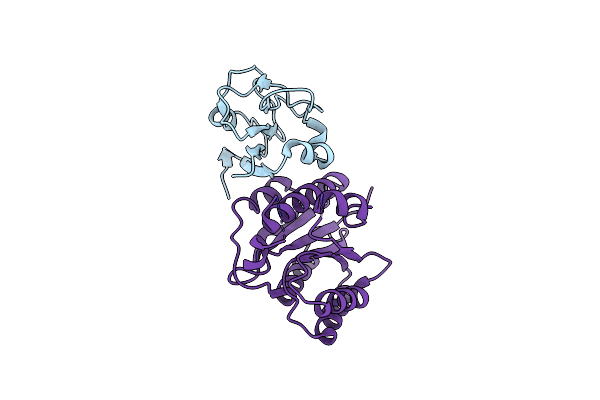
Crystal Structure Of Binary Complex Between Aster Yellows Witches'-Broom Phytoplasma Effector Sap05 And The Zinc Finger Domain Of Spl5 From Arabidopsis Thaliana
Organism: Aster yellows witches'-broom phytoplasma aywb, Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-07-05 Classification: PROTEIN BINDING Ligands: ZN |
 |
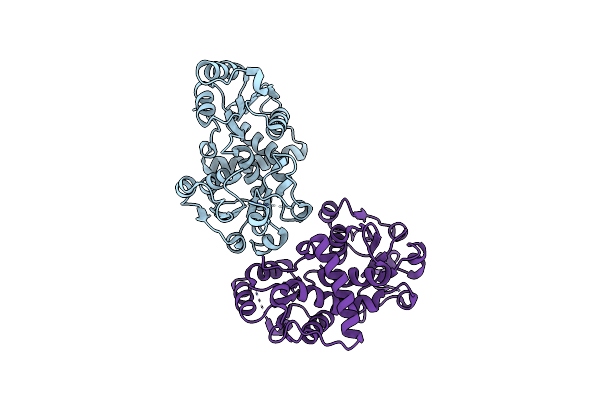
Crystal Structure Of Binary Complex Between Aster Yellows Witches'-Broom Phytoplasma Effector Sap05 And The Von Willebrand Factor Type A Domain Of The Proteasomal Ubiquitin Receptor Rpn10 From Arabidopsis Thaliana
Organism: Aster yellows witches'-broom phytoplasma aywb, Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2023-07-05 Classification: PROTEIN BINDING |
 |
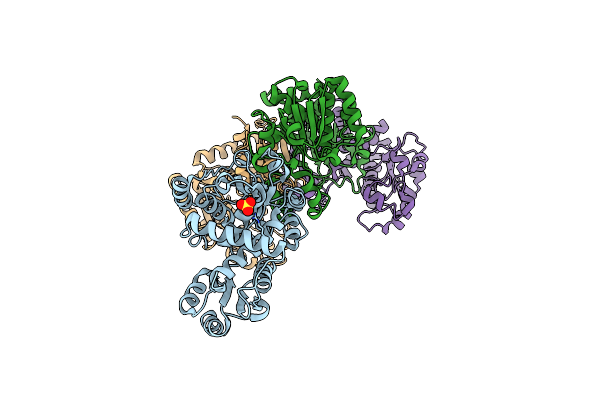
Anabolic Ornithine Carbamoyltransferases (Otcs) From Psychrobacter Sp. Pamc 21119
Organism: Psychrobacter sp. pamc 21119
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-08-31 Classification: TRANSFERASE |
 |
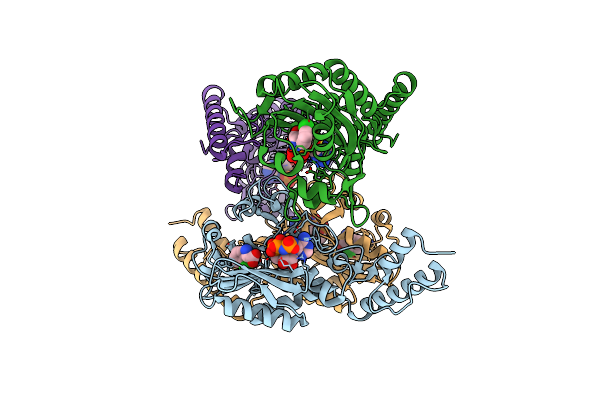
Catabolic Ornithine Carbamoyltransferases (Otcs) From Psychrobacter Sp. Pamc 21119
Organism: Psychrobacter sp. pamc 21119
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-08-31 Classification: TRANSFERASE Ligands: SO4 |
 |
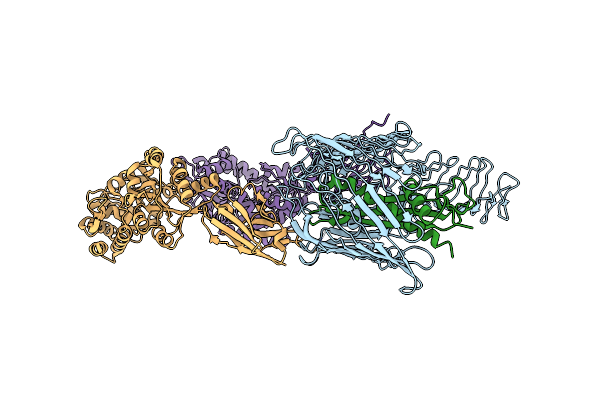
Crystal Structure Of Pyrrolysyl-Trna Synthetase (N346D/C348S/Y384F) In Complex With O-Chlorophenylalanine And Amp-Pnp
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-07-20 Classification: RNA BINDING PROTEIN Ligands: ANP, 2L5 |
 |
Cryo-Em Structure Of N. Gonorhoeae Lptde In Complex With Promacrobodies (Mbps Have Not Been Built De Novo)
Organism: Neisseria gonorrhoeae, Synthetic construct, Methanosarcina mazei
Method: ELECTRON MICROSCOPY Release Date: 2022-05-04 Classification: TRANSPORT PROTEIN |
 |
Re-Refinement Of The 2Xry X-Ray Structure Of Archaeal Class Ii Cpd Photolyase From Methanosarcina Mazei
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-03-09 Classification: LYASE Ligands: SO4, GOL, FAD |

