Search Count: 832
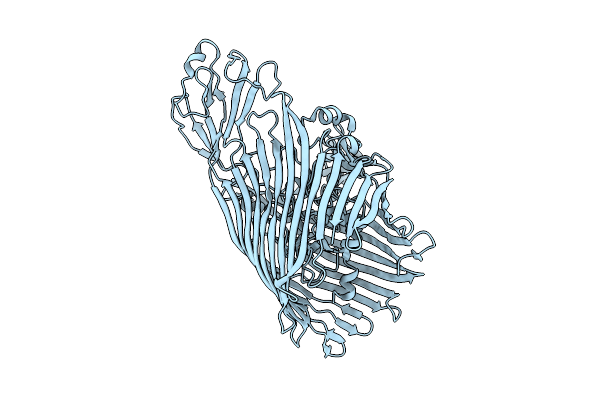 |
Fusa (Ferredoxin Receptor From Pectobacterium Atrosepticum) In The Presence Of Ra-Lps
Organism: Pectobacterium atrosepticum scri1043
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of D-Cysteine Desulfhydrase From Pectobacterium Atrosepticum
Organism: Pectobacterium atrosepticum scri1043
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-12-14 Classification: LYASE |
 |
Crystal Structure Of D-Cysteine Desulfhydrase With A Trapped Plp-Pyruvate Geminal Diamine
Organism: Pectobacterium atrosepticum scri1043
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2022-12-14 Classification: LYASE Ligands: FMT, EDO |
 |
Structure Of The Ligand Binding Domain Of The Paca (Eca2226) Chemoreceptor Of Pectobacterium Atrosepticum Scri1043 In Complex With Betaine.
Organism: Pectobacterium atrosepticum scri1043
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2022-05-11 Classification: SIGNALING PROTEIN Ligands: BET, GOL |
 |
Organism: Pectobacterium atrosepticum (strain scri 1043 / atcc baa-672)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-02-17 Classification: HYDROLASE Ligands: ACT, NA, GOL, SO4 |
 |
The Crystal Structure Of The Ferredoxin Protease Fusc In Complex With Its Substrate Plant Ferredoxin
Organism: Pectobacterium atrosepticum scri1043, Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-06-20 Classification: HYDROLASE |
 |
The Crystal Structure Of The Ferredoxin Protease Fusc E83A Mutant In Complex With Arabidopsis Ferredoxin
Organism: Pectobacterium atrosepticum (strain scri 1043 / atcc baa-672), Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-06-20 Classification: HYDROLASE Ligands: ZN |
 |
The Crystal Structure Of The Ferredoxin Protease Fusc In Complex With Arabidopsis Ferredoxin, Ethylmercury Phosphate Soaked Dataset
Organism: Pectobacterium atrosepticum (strain scri 1043 / atcc baa-672), Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-06-20 Classification: HYDROLASE Ligands: PO4, HG |
 |
The Crystal Structure Of The Ferredoxin Receptor Fusa From Pectobacterium Atrosepticum Scri1043
Organism: Pectobacterium atrosepticum
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-08-31 Classification: TRANSPORT PROTEIN Ligands: LDA, BOG |
 |
Organism: Pectobacterium atrosepticum (strain scri 1043 / atcc baa-672)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-03-16 Classification: DNA BINDING PROTEIN |
 |
Organism: Pectobacterium atrosepticum scri1043
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-10-07 Classification: HYDROLASE |
 |
Ec-Nmr Structure Of Erwinia Carotovora Eca1580 N-Terminal Domain Determined By Combining Evolutionary Couplings (Ec) And Sparse Nmr Data. Northeast Structural Genomics Consortium Target Ewr156A
Organism: Pectobacterium atrosepticum scri1043
Method: SOLUTION NMR Release Date: 2015-07-01 Classification: Structural Genomics, Unknown Function |
 |
Crystal Structure Of A Domain Of Unknown Function (Duf1537) From Pectobacterium Atrosepticum (Eca3761), Target Efi-511609, With Bound D-Threonate, Domain Swapped Dimer
Organism: Pectobacterium atrosepticum
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2015-04-01 Classification: Structural Genomics, Unknown function Ligands: THE |
 |
Crystal Structure Of A Domain Of Unknown Function (Duf1537) From Pectobacterium Atrosepticum (Eca3761), Target Efi-511609, Apo Structure, Domain Swapped Dimer
Organism: Pectobacterium atrosepticum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-02-18 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
 |
Crystal Structure Of Periplasmic Solute Binding Protein Eca2210 From Pectobacterium Atrosepticum Scri1043, Target Efi-510858
Organism: Pectobacterium atrosepticum
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2014-12-24 Classification: TRANSPORT PROTEIN Ligands: GOL, ACT |
 |
Crystal Structure Of The Octaprenyl-Methyl-Methoxy-Benzq Molecule From Erwina Carotovora Subsp. Atroseptica Strain Scri 1043 / Atcc Baa-672, Northeast Structural Genomics Consortium (Nesg) Target Ewr161
Organism: Pectobacterium atrosepticum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-11-20 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Eca2234 Protein From Erwinia Carotovora, Northeast Structural Genomics Consortium Target Ewr44
Organism: Pectobacterium atrosepticum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-04-14 Classification: structural genomics, unknown function |
 |
Crystal Structure Of Putative Sugar Isomerase (Yp_050048.1) From Erwinia Carotovora Atroseptica Scri1043 At 1.54 A Resolution
Organism: Pectobacterium atrosepticum scri1043
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2009-12-22 Classification: ISOMERASE Ligands: FE, MG, CL, UNL, EDO |
 |
Crystal Structure Of A Putative Delta-5-3-Ketosteroid Isomerase (Eca2236) From Pectobacterium Atrosepticum Scri1043 At 1.55 A Resolution
Organism: Pectobacterium atrosepticum scri1043
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2008-12-09 Classification: ISOMERASE |
 |
Crystal Structure Of A Protein Of Unknown Function (Eca1910) From Pectobacterium Atrosepticum Scri1043 At 2.20 A Resolution
Organism: Pectobacterium atrosepticum scri1043
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-04-01 Classification: UNKNOWN FUNCTION Ligands: EDO |

