Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Crystal Structure Of A Calcium Bound C2 Domain Containing Protein From Trichomonas Vaginalis
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: 1PE, CL, PEG, PGE |
 |
Crystal Structure Of A Calcium Bound C2 Domain Containing Protein From Trichomonas Vaginalis (P21 Form)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: PG4, CA, MES, 1PE |
 |
Crystal Structure Of A Calcium Bound C2 Domain Containing Protein From Trichomonas Vaginalis (Orthorhombic P Form)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: CA, 1PE |
 |
Crystal Structure Of A C2 Domain Containing Protein From Trichomonas Vaginalis
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: GOL, PGE |
 |
Crystal Structure Of A Malonate Bound C2 Domain Containing Protein From Trichomonas Vaginalis (P21 Form)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: MLI, CL, NA |
 |
Crystal Structure Of A Calcium Bound C2 Domain Containing Protein From Trichomonas Vaginalis (P61 Form)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: CA, CL, PG4 |
 |
Crystal Structure Of A C2 Domain Containing Protein From Trichomonas Vaginalis In Complex With Pyrophosphate
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: LIPID BINDING PROTEIN Ligands: PPV |
 |
Crystal Structure Of Phosphoglycerate Mutase From Trichomonas Vaginalis In Complex With 3-Phosphoglyceric Acid
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: ISOMERASE Ligands: CL, 3PG, PG4, SO4 |
 |
Organism: Paenibacillus sp. fsl e2-0178
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: RNA Ligands: GUN, MG |
 |
Crystal Structure Of Guanine-Ii Riboswitch In Complex With 2'-Deoxyguanosine
Organism: Paenibacillus sp. fsl e2-0178
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: RNA Ligands: MG, GNG |
 |
Organism: Paenibacillus sp. fsl e2-0178
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: RNA Ligands: HPA, GTP, MG |
 |
Organism: Paenibacillus sp. fsl e2-0178
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: RNA Ligands: GMP, MG |
 |
Crystal Structure Of Guanine-Ii Riboswitch In Complex With Guanine Soaked With Mn2+
Organism: Paenibacillus sp. fsl e2-0178
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: RNA Ligands: GUN, MN, GTP, MG |
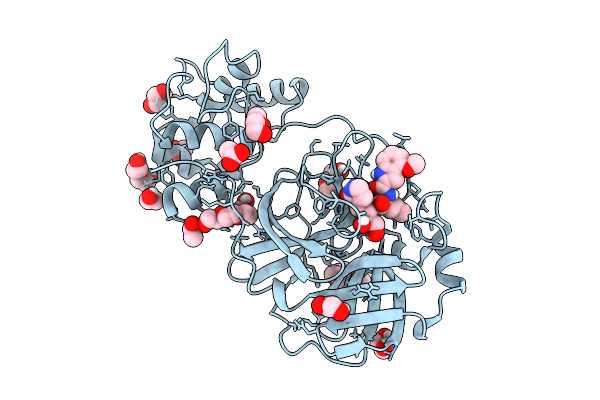 |
Co-Crystal Structure Of Feline Coronavirus Uu23 Main Protease With Pfizer Intravenous Compound Pf-00835231
Organism: Feline alphacoronavirus 1
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN Ligands: V2M, ACY, PG4, EDO, PEG |
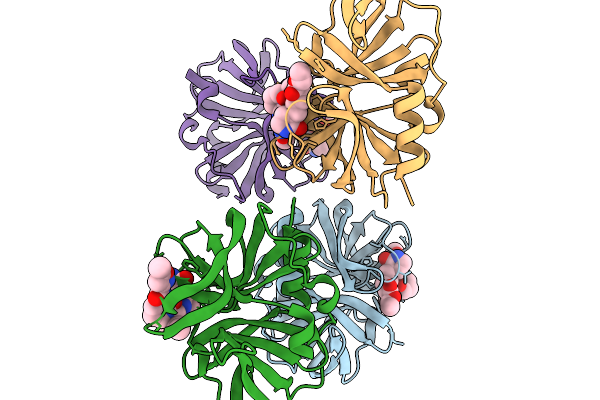 |
Organism: Rhinovirus b14
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: HYDROLASE Ligands: XNV |
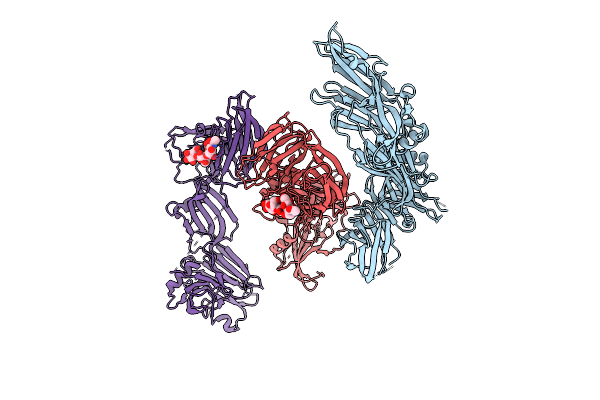 |
Crystal Structure Of Ha3 From Clostridium Botulinum Type B With Alpha2,3-Sialyllactose
Organism: Clostridium botulinum b1 str. okra
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TOXIN |
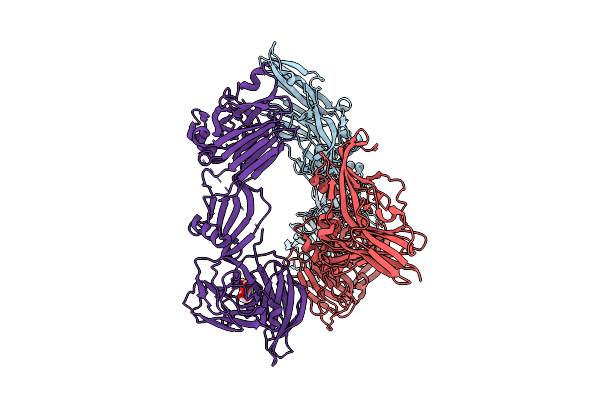 |
Crystal Structure Of Ha3 From Clostridium Botulinum Type B With Alpha2,6-Sialyllactose
Organism: Clostridium botulinum b1 str. okra
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TOXIN Ligands: SIA |
 |
Crystal Structure Of Phosphoglycerate Mutase From Trichomonas Vaginalis (Sulfate Bound)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2025-04-23 Classification: ISOMERASE Ligands: SO4 |
 |
Organism: Flavobacteriales bacterium
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2025-04-16 Classification: OXIDOREDUCTASE Ligands: GOL, HEM, CTE |
 |
Organism: Flavobacteriales bacterium
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2025-04-16 Classification: OXIDOREDUCTASE Ligands: GOL, HEM, TRP, TRS |