Search Count: 410
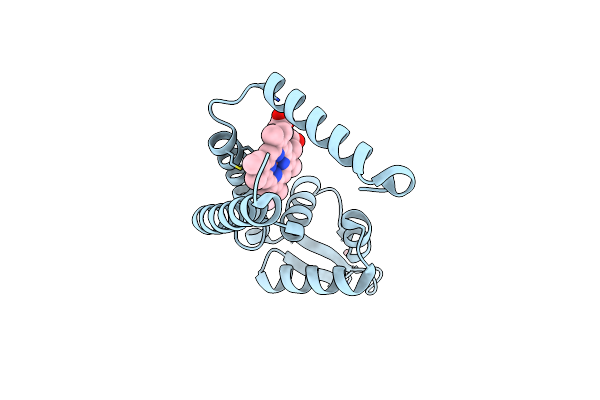 |
Crystal Structure Of Heme Sensor Protein Pefr From Streptococcus Agalactiae In Complex With Heme
Organism: Streptococcus agalactiae serotype iii (strain nem316)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-09-29 Classification: TRANSCRIPTION Ligands: HEM, CO |
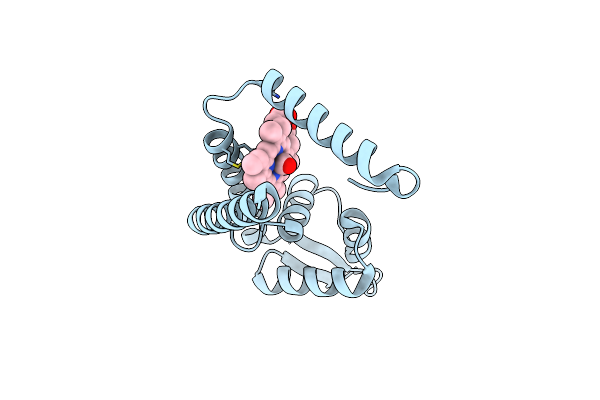 |
Crystal Structure Of Heme Sensor Protein Pefr In Complex With Heme And Carbon Monoxide
Organism: Streptococcus agalactiae serotype iii (strain nem316)
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2021-09-29 Classification: TRANSCRIPTION Ligands: HEM, CMO |
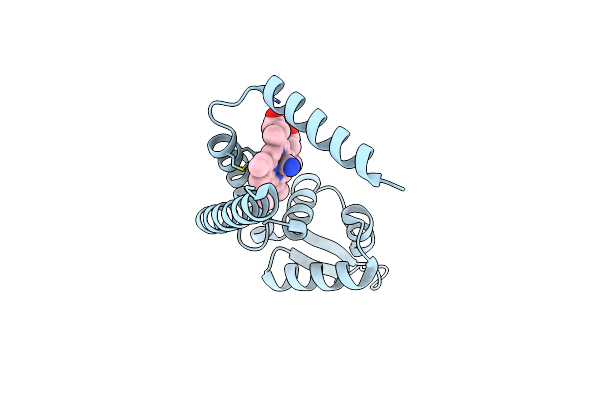 |
Crystal Structure Of Heme Sensor Protein Pefr In Complex With Heme And Cyanide
Organism: Streptococcus agalactiae serotype iii (strain nem316)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-09-29 Classification: TRANSCRIPTION Ligands: HEM, CYN |
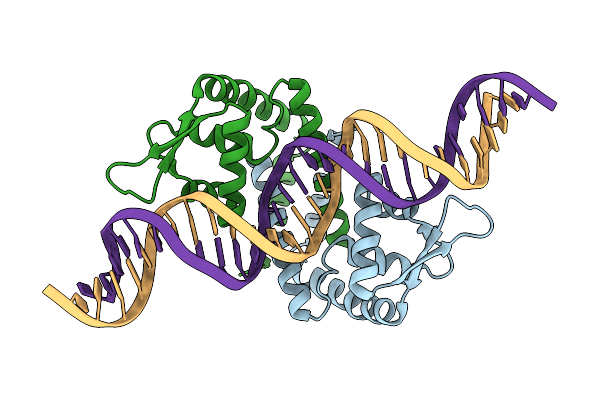 |
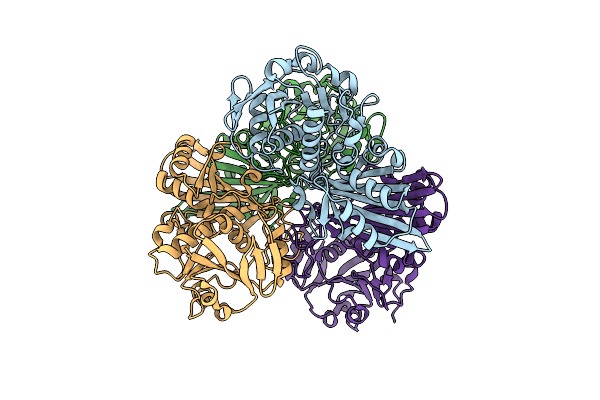
Heme Sensor Protein Pefr From Streptococcus Agalactiae Bound To Operator Dna (28-Mer)
Organism: Streptococcus agalactiae serotype iii (strain nem316), Streptococcus agalactiae nem316
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2021-09-29 Classification: TRANSCRIPTION |
 |
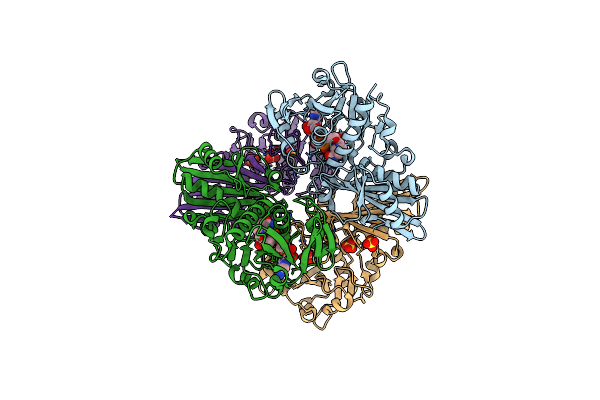
Organism: Streptococcus agalactiae nem316
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-01-02 Classification: CELL ADHESION |
 |
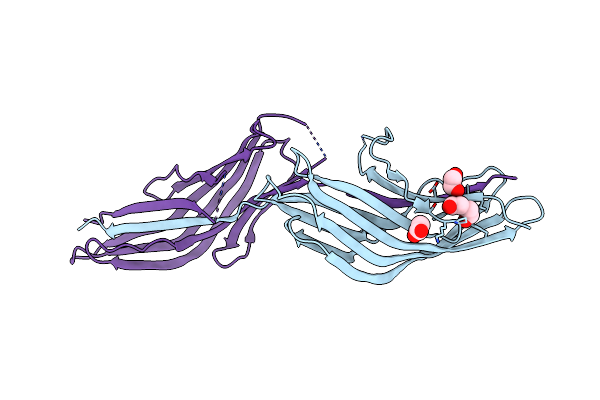
Organism: Streptococcus agalactiae serotype iii (strain nem316)
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2018-04-18 Classification: TRANSFERASE Ligands: NAD, SO4 |
 |
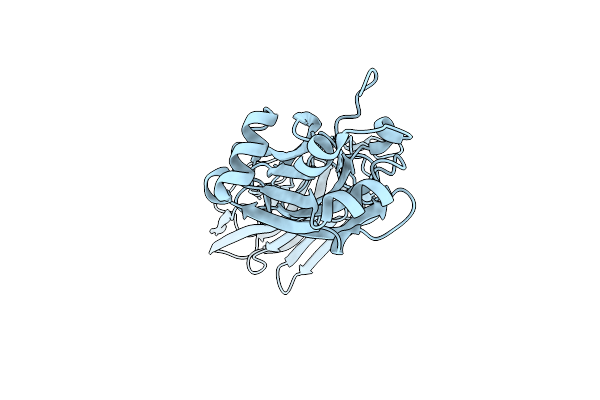
Organism: Streptococcus agalactiae serotype iii (strain nem316)
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2016-06-22 Classification: CELL ADHESION Ligands: ACT, PEG, EDO |
 |
Organism: Streptococcus agalactiae serotype iii (strain nem316)
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2016-06-22 Classification: CELL ADHESION |
 |
Organism: Streptococcus agalactiae serotype iii (strain nem316)
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2016-06-22 Classification: CELL ADHESION Ligands: EDO, PEG |
 |
Crystal Structure Of Keratin 4 Binding Domain Of Surface Adhesin Srr-1 Of S.Agalactiae
Organism: Streptococcus agalactiae nem316
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-02-11 Classification: CELL ADHESION |
 |
Crystal Structure Of 4-Deoxy-L-Threo-5-Hexosulose-Uronate Ketol-Isomerase From Streptococcus Agalactiae
Organism: Streptococcus agalactiae nem316
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-01-21 Classification: ISOMERASE |
 |
Crystal Structure Of 4-Deoxy-L-Threo-5-Hexosulose-Uronate Ketol-Isomerase Complexed With A Tartrate
Organism: Streptococcus agalactiae nem316
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2014-12-24 Classification: ISOMERASE Ligands: TLA |
 |
Crystal Structure Of 2-Keto-3-Deoxy-D-Gluconate Dehydrogenase From Streptococcus Agalactiae
Organism: Streptococcus agalactiae nem316
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2014-12-24 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Glyceraldehyde-3-Phosphate Dehydrogenase From Streptococcus Agalactiae Nem316 At 2.46 Angstrom Resolution
Organism: Streptococcus agalactiae
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2014-10-15 Classification: OXIDOREDUCTASE Ligands: NAD, EDO |
 |
Crystal Structure Of Phosphotransferase System Component Eiia From Streptococcus Agalactiae
Organism: Streptococcus agalactiae nem316
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-08-20 Classification: TRANSFERASE Ligands: GOL |
 |
Crystal Structure Of Unsaturated Glucuronyl Hydrolase Mutant D115N/K370S From Streptococcus Agalactiae
Organism: Streptococcus agalactiae
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2014-05-28 Classification: HYDROLASE Ligands: EDO |
 |
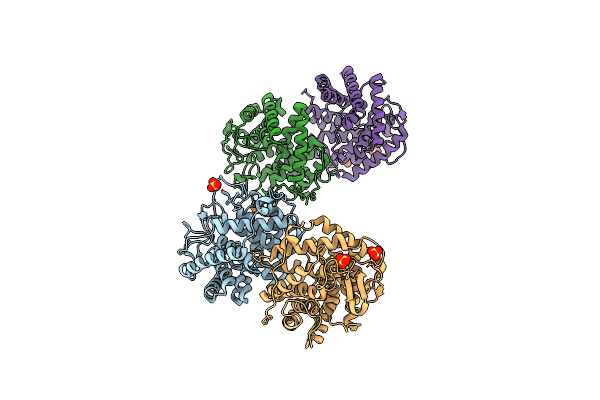
1.65 Angstrom Crystal Structure Of Serine-Rich Repeat Adhesion Glycoprotein (Srr1) From Streptococcus Agalactiae
Organism: Streptococcus agalactiae
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2013-11-06 Classification: PROTEIN BINDING Ligands: CA, BME, MES |
 |
Crystal Structure Of Unsaturated Glucuronyl Hydrolase Mutant D115N From Streptcoccus Agalactiae
Organism: Streptococcus agalactiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-10-03 Classification: HYDROLASE Ligands: SO4 |
 |
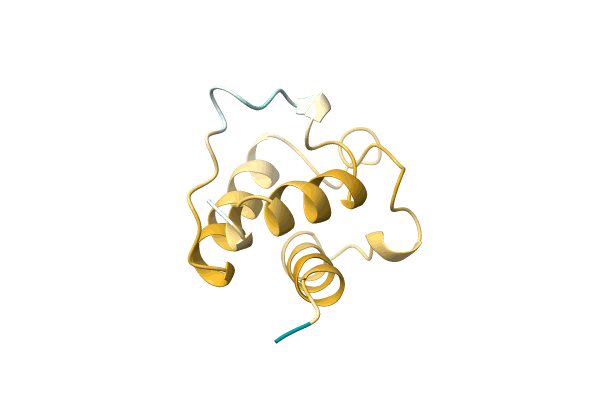
Crystal Structure Of Gbs1074, An Esat-6 Homologue From Group B Streptococcus
Organism: Streptococcus agalactiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-11-10 Classification: UNKNOWN FUNCTION |
 |
NA
Organism: Streptococcus agalactiae serotype III (strain NEM316)
Method: Alphafold Release Date: Classification: NA Ligands: NA |

