Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
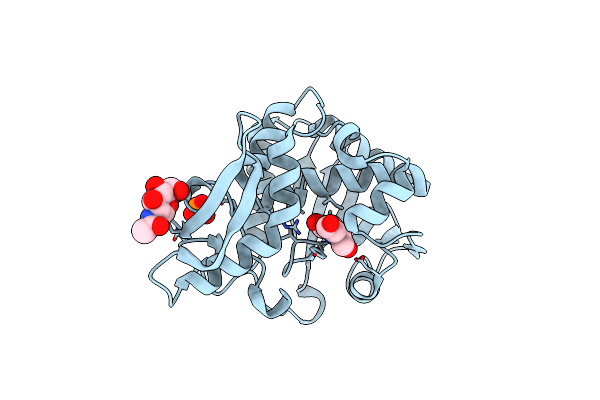
Cs H-Nox Mutant With Unnatural Amino Acid 4-Cyano-L-Phenylalanine At Site 5
Organism: Caldanaerobacter subterraneus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2018-05-16 Classification: OXYGEN BINDING Ligands: HEM, 211 |
 |
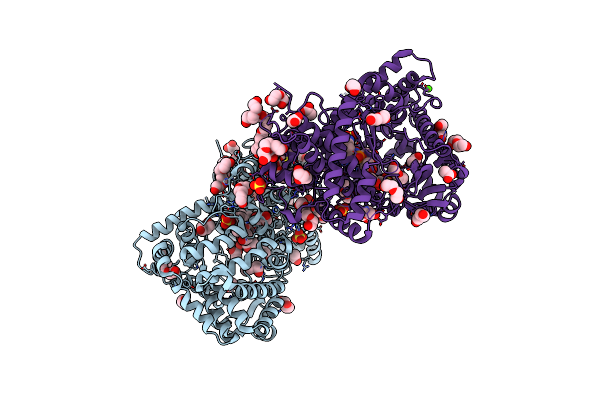
Structure Of The Complex Of Type 1 Ribosome Inactivating Protein With Triethanolamine At 1.88 Angstrom Resolution
Organism: Momordica balsamina
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2015-08-12 Classification: SIGNALING PROTEIN Ligands: NAG, 211, PO4 |
 |
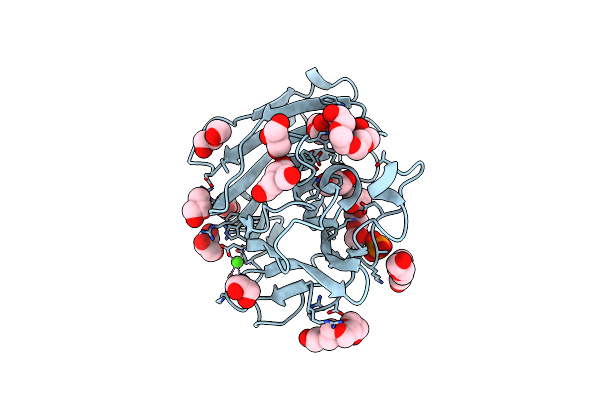
Crystal Structure Of Mycobacterium Tuberculosis Zinc Metalloprotease Zmp1 In Complex With Inhibitor
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-08-03 Classification: HYDROLASE/INHIBITOR Ligands: ZN, RDF, 211, PEG, PGE, PG4, ACT, SO4, CL, CA |
 |
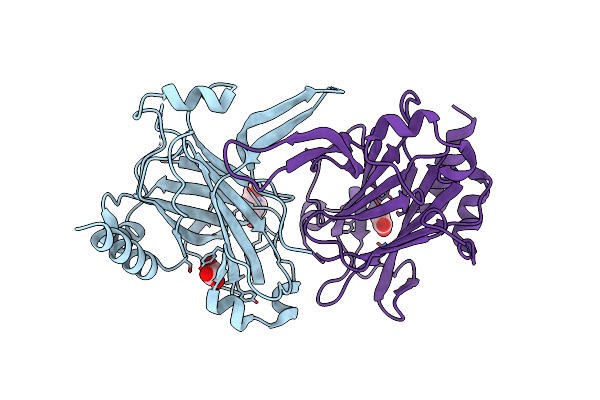
The Crystal Structure Of A Glycosyl Hydrolases (Gh) Family Protein 16 From Mycobacterium Smegmatis Str. Mc2 155
Organism: Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2011-05-11 Classification: HYDROLASE Ligands: CA, PO4, PEG, PG4, GOL, 211 |
 |
The 2.25 A Crystal Structure Of Lipl32, The Major Surface Antigen Of Leptospira Interrogans Serovar Copenhageni
Organism: Leptospira interrogans serovar copenhageni
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2009-06-16 Classification: MEMBRANE PROTEIN Ligands: 211, OXL, CL |
 |
Crystal Structure Of Arabidopsis Thaliana Agmatine Deiminase From Cell Free Expression
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-05-26 Classification: HYDROLASE Ligands: CL, MG, NA, K, 211, 1PE |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-09-19 Classification: STRUCTURAL PROTEIN Ligands: SR, SPD, ATP, EDO, 211 |