Search Count: 334
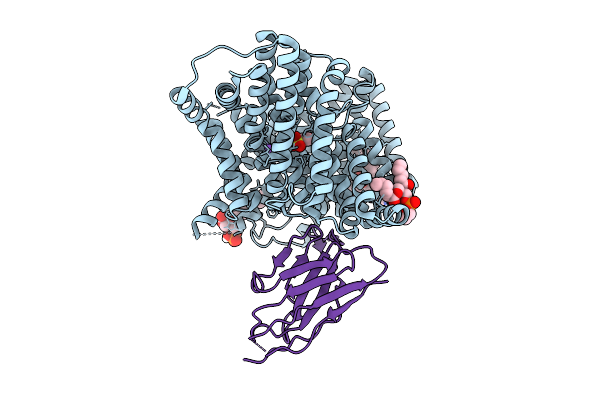 |
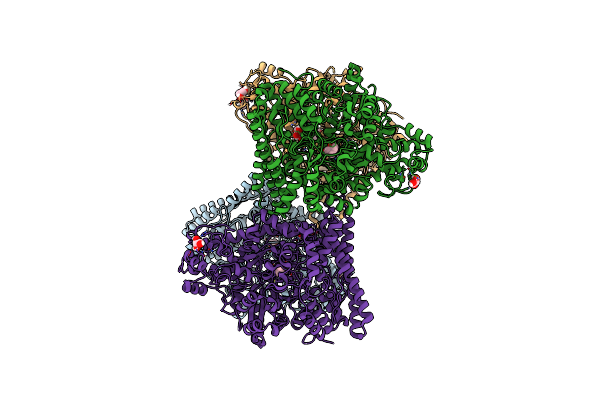
Cryo-Em Structure Of The Isethionate Trap Transporter Iseqm From Oleidesulfovibrio Alaskensis With Bound Isethionate
Organism: Oleidesulfovibrio alaskensis g20, Helicobacter pylori
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: TRANSPORT PROTEIN |
 |
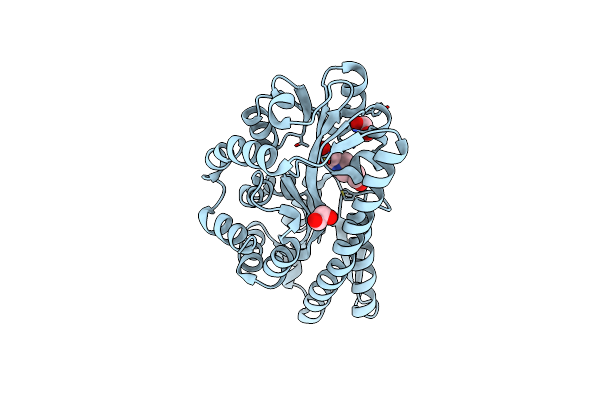
Crystal Structure Of A Mes Bound Substrate Binding Protein (Isep) From An Isethionate Trap Transporter
Organism: Oleidesulfovibrio alaskensis g20
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-08-28 Classification: TRANSPORT PROTEIN Ligands: MES |
 |
Organism: Oleidesulfovibrio alaskensis g20
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2024-08-28 Classification: TRANSPORT PROTEIN Ligands: EPE, EDO |
 |
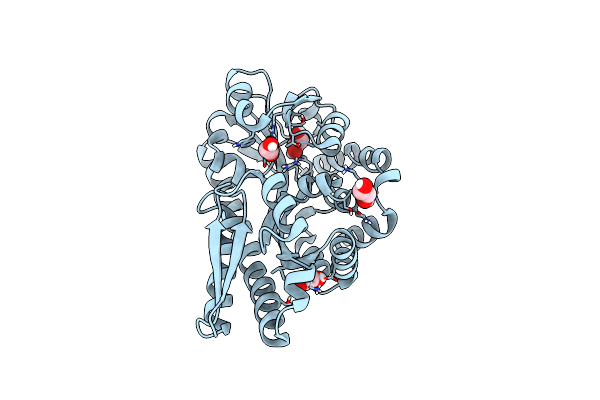
Crystal Structure Of An Isethionate Bound Substrate Binding Protein (Isep) From An Isethionate Trap Transporter
Organism: Oleidesulfovibrio alaskensis g20
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2024-07-10 Classification: TRANSPORT PROTEIN Ligands: 8X3, NO3, EDO |
 |
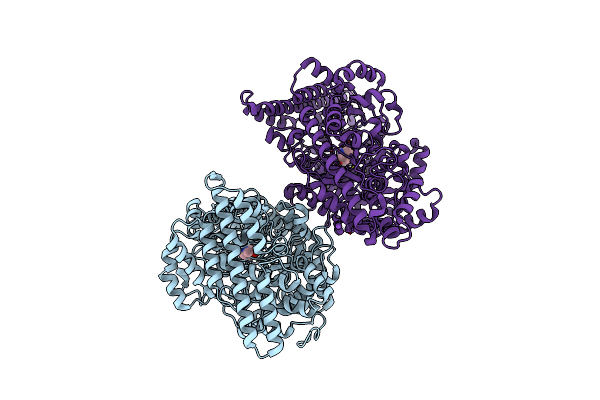
Apo Crystal Structure Of A Substrate Binding Protein (Isep) From An Isethionate Trap Transporter
Organism: Oleidesulfovibrio alaskensis g20
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2024-06-26 Classification: TRANSPORT PROTEIN Ligands: EDO, CL |
 |
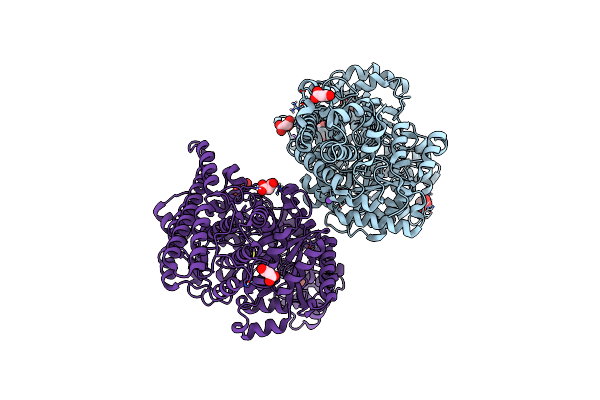
Wild-Type Choline Tma Lyase In Complex With 1-Methyl-1,2,3,6-Tetrahydropyridin-3-Ol
Organism: Desulfovibrio alaskensis (strain g20)
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2020-11-04 Classification: LYASE/INHIBITOR |
 |
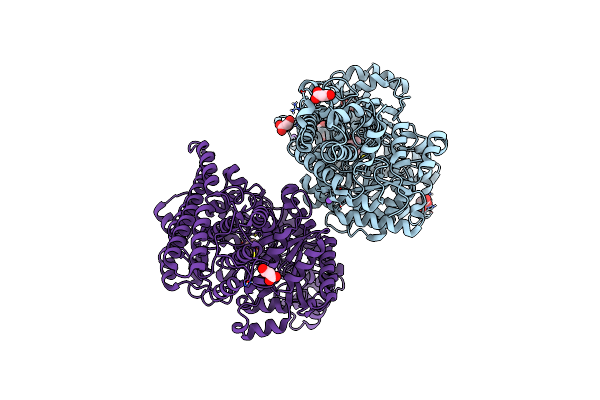
Organism: Desulfovibrio alaskensis (strain g20)
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2019-01-16 Classification: LYASE Ligands: BTL |
 |
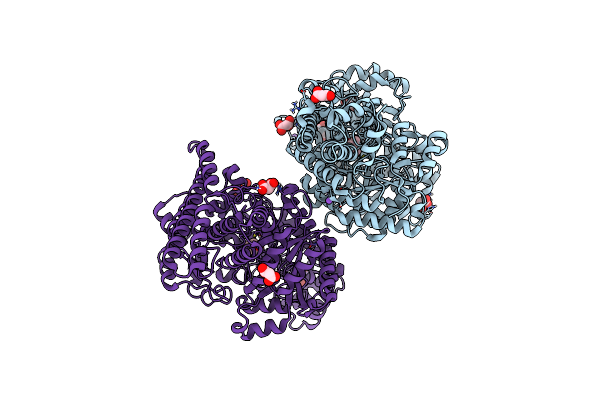
Organism: Desulfovibrio alaskensis (strain g20)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-08-02 Classification: TRANSPORT PROTEIN |
 |
Organism: Desulfovibrio alaskensis (strain g20)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-09-28 Classification: LYASE Ligands: CHT, GOL |
 |
Organism: Desulfovibrio alaskensis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-09-28 Classification: LYASE |
 |
Organism: Desulfovibrio alaskensis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-09-28 Classification: LYASE |
 |
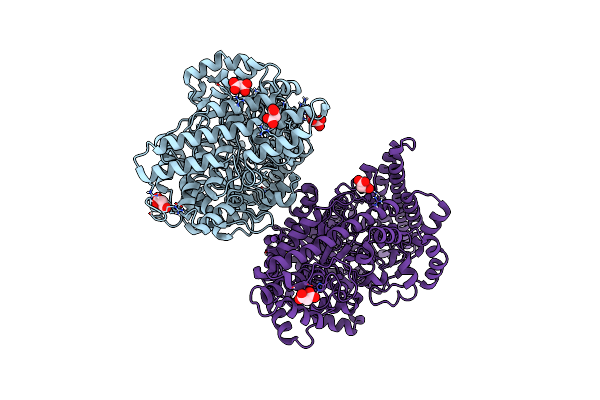
Organism: Desulfovibrio alaskensis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-09-28 Classification: LYASE |
 |
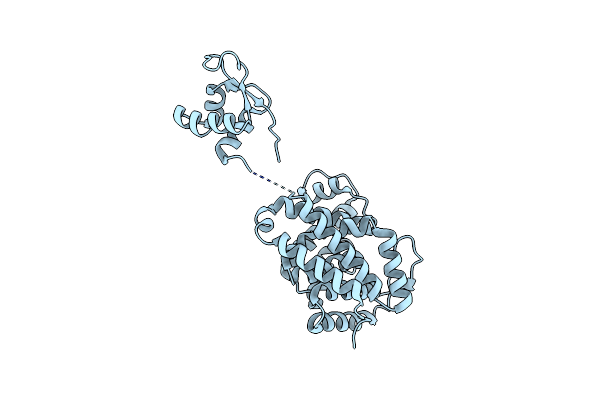
Organism: Desulfovibrio alaskensis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-09-28 Classification: LYASE |
 |
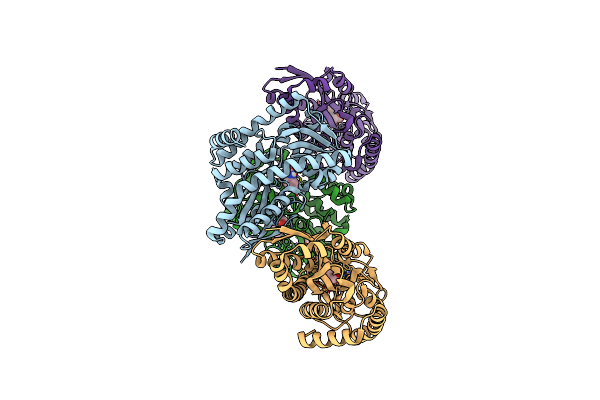
Crystal Structure Of A Fic Family Protein (Dde_2494) From Desulfovibrio Desulfuricans G20 At 2.70 A Resolution
Organism: Desulfovibrio alaskensis
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2014-10-29 Classification: DNA BINDING PROTEIN Ligands: UNL |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Desulfovibrio Alaskensis G20 (Dde_0634, Target Efi-510120) With Bound 3-Indole Acetic Acid
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2014-06-18 Classification: SOLUTE-BINDING PROTEIN Ligands: IAC, EDO |
 |
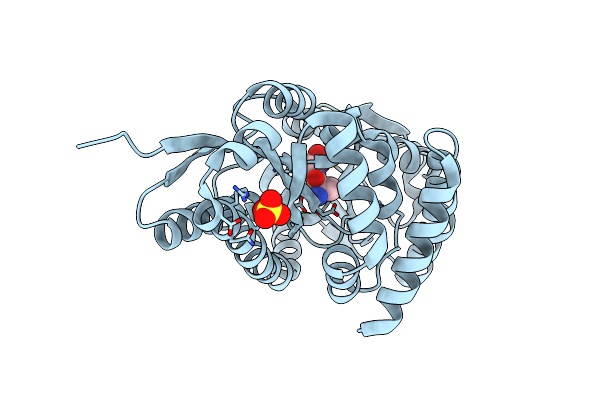
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Desulfovibrio Alaskensis G20 (Dde_0634, Target Efi-510120) With Bound Indole Pyruvate
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-05-28 Classification: TRANSPORT PROTEIN Ligands: 3IO, EDO |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Desulfovibrio Alaskensis G20 (Dde_1548), Target Efi-510103, With Bound D-Ala-D-Ala
Organism: Desulfovibrio alaskensis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-12-04 Classification: TRANSPORT PROTEIN Ligands: CL, DAL, SO4 |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Desulfovibrio Alaskensis G20 (Dde_0634), Target Efi-510102, With Bound D-Tryptophan
Organism: Desulfovibrio alaskensis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-11-13 Classification: TRANSPORT PROTEIN Ligands: DTR |
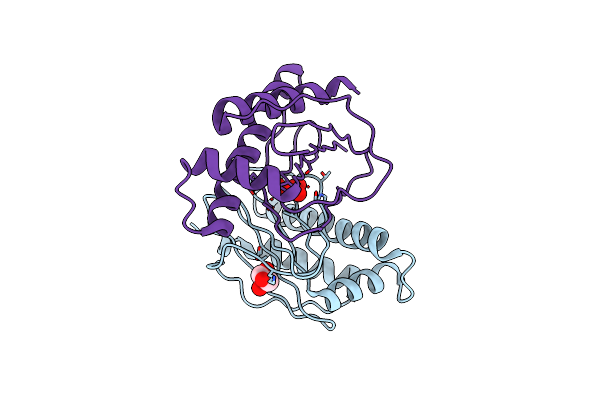 |
Crystal Structure Of A Chemotaxis Protein Chex (Dde_0281) From Desulfovibrio Desulfuricans Subsp. At 1.30 A Resolution
Organism: Desulfovibrio desulfuricans subsp. desulfuricans str. g20
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2009-06-23 Classification: HYDROLASE Ligands: UNL, GOL |
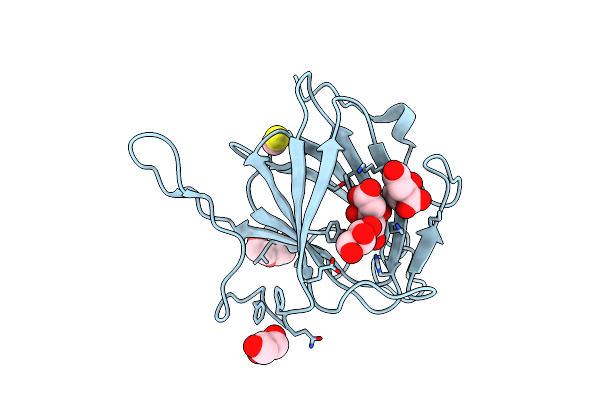 |
Crystal Structure Of Dtdp Sugar Isomerase (Yp_390184.1) From Desulfovibrio Desulfuricans G20 At 1.95 A Resolution
Organism: Desulfovibrio desulfuricans subsp. desulfuricans str. g20
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2008-09-30 Classification: ISOMERASE Ligands: UNL, CIT, PG4, GOL |

