Search Count: 30
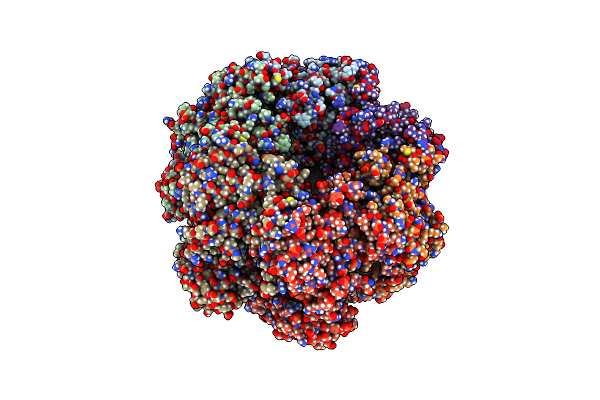 |
Organism: Candidatus mancarchaeum acidiphilum
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2025-03-12 Classification: REPLICATION Ligands: ADP, PO4, ZN |
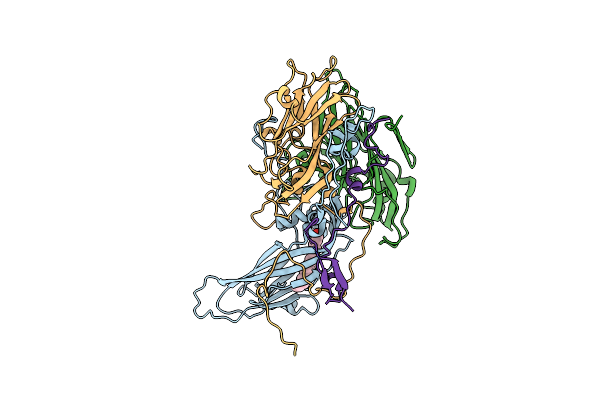 |
Structure Of Coxsackievirus B5 Capsid (Mutant Cvb5F.Cas.Genogroupb) - F Particle
Organism: Coxsackievirus b5
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: VIRUS Ligands: PLM |
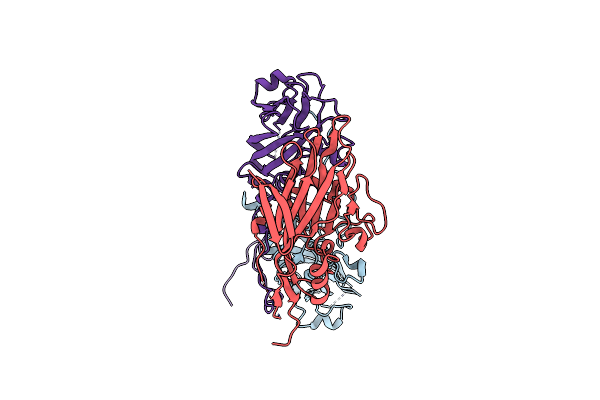 |
Structure Of Coxsackievirus B5 Capsid (Mutant Cvb5F.Cas.Genogroupb) - A Particle
Organism: Coxsackievirus b5
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: VIRUS |
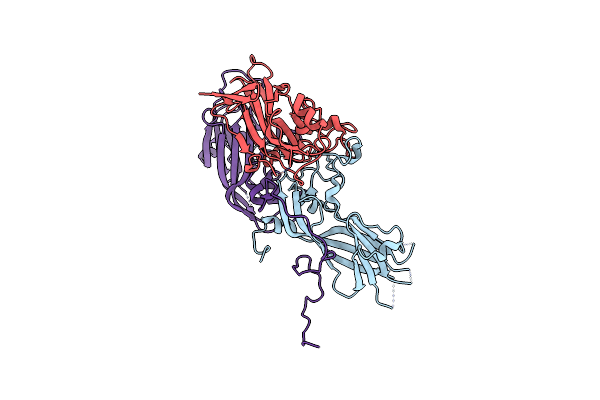 |
Structure Of Coxsackievirus B5 Capsid (Mutant Cvb5F.Cas.Genogroupb) - E Particle
Organism: Coxsackievirus b5
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: VIRUS |
 |
Organism: Coxsackievirus b5
Method: ELECTRON MICROSCOPY Release Date: 2022-03-30 Classification: VIRUS |
 |
Organism: Pseudonocardia autotrophica
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-05-17 Classification: OXIDOREDUCTASE Ligands: HEM, PG0 |
 |
Organism: Pseudonocardia autotrophica
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-05-17 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Organism: Streptococcus pyogenes m1 476
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2016-01-13 Classification: UNKNOWN FUNCTION Ligands: GOL |
 |
Organism: Pseudonocardia autotrophica
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2013-04-17 Classification: OXIDOREDUCTASE Ligands: HEM, VD3 |
 |
Structure Of Cytochrome P450 Vdh Mutant (Vdh-K1) Obtained By Directed Evolution
Organism: Pseudonocardia autotrophica
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-07-28 Classification: OXIDOREDUCTASE Ligands: HEM, CA, ACT, GOL |
 |
Structure Of Cytochrome P450 Vdh Mutant (Vdh-K1) Obtained By Directed Evolution With Bound Vitamin D3
Organism: Pseudonocardia autotrophica
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2010-07-28 Classification: OXIDOREDUCTASE Ligands: HEM, VD3, ACT, CA, GOL |
 |
Structure Of Cytochrome P450 Vdh Mutant (Vdh-K1) Obtained By Directed Evolution With Bound 25-Hydroxyvitamin D3
Organism: Pseudonocardia autotrophica
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-07-28 Classification: OXIDOREDUCTASE Ligands: HEM, VDY, CA, GOL, ACT |
 |
Structure Of Cytochrome P450 Vdh From Pseudonocardia Autotrophica (Trigonal Crystal Form)
Organism: Pseudonocardia autotrophica
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2010-07-14 Classification: OXIDOREDUCTASE Ligands: HEM, PG0 |
 |
Structure Of Cytochrome P450 Vdh From Pseudonocardia Autotrophica (Orthorhombic Crystal Form)
Organism: Pseudonocardia autotrophica
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2010-07-14 Classification: OXIDOREDUCTASE Ligands: HEM, CA |
 |
Organism: Depressariinae sp. LA2011_162
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Hartmannibacter diazotrophicus
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Hartmannibacter diazotrophicus
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Hartmannibacter diazotrophicus
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
 |

