Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
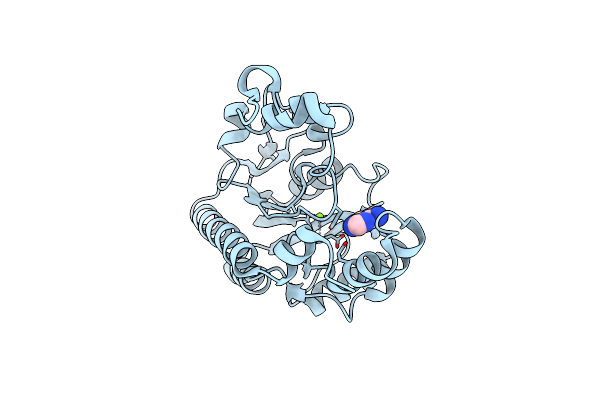 |
Crystal Structure Of Non-Specific Class-C Acid Phosphatase From Sphingobium Sp. Rsms Bound To Adenine At Ph 9
Organism: Sphingobium sp. 20006fa
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-07-06 Classification: HYDROLASE Ligands: ADE, MG |
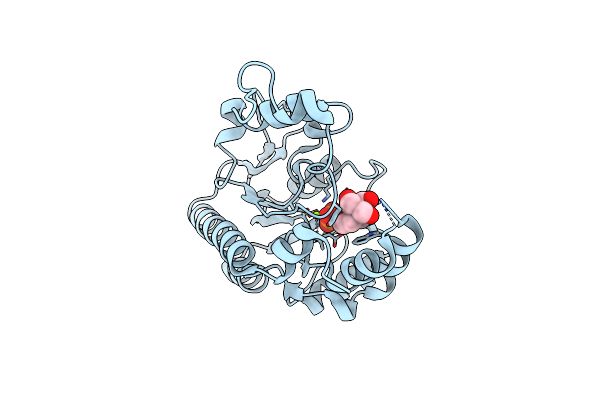 |
Crystal Structure Of Non-Specific Class-C Acid Phosphatase From Sphingobium Sp. Rsms Bound To Bis-Tris At Ph 5.5
Organism: Sphingobium sp. 20006fa
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2022-07-06 Classification: HYDROLASE Ligands: BTB, MG, PO4 |
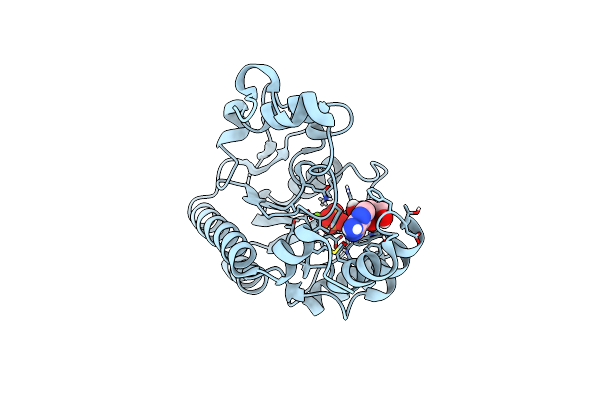 |
Crystal Structure Of Non-Specific Class-C Acid Phosphatase From Sphingobium Sp. Rsms Bound To Adenosine At Ph 5.5
Organism: Sphingobium sp. 20006fa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-07-06 Classification: HYDROLASE Ligands: ADN, PO4, MG |
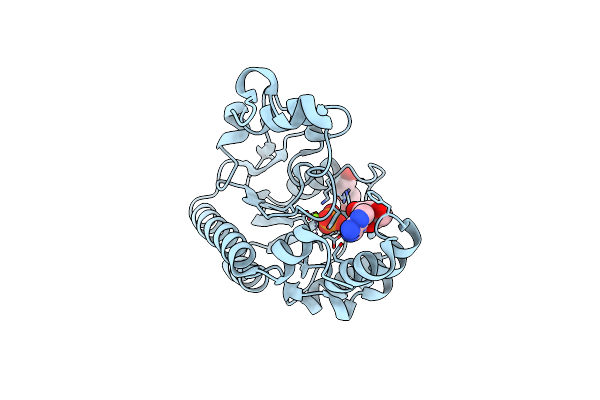 |
Crystal Structure Of Non-Specific Class-C Acid Phosphatase From Sphingobium Sp. Rsms Bound To Adenosine At Ph 5.5
Organism: Sphingobium sp. 20006fa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-07-06 Classification: HYDROLASE Ligands: ADN, PEG, MG, PO4 |
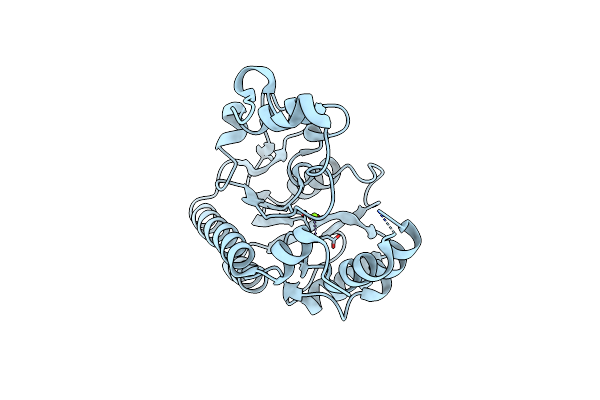 |
Organism: Sphingobium sp. 20006fa
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2021-11-10 Classification: HYDROLASE Ligands: MG |