Search Count: 437
 |
Organism: Parachlamydia sp., Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: TLA |
 |
Organism: Homo sapiens, Parachlamydia sp.
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: CIT |
 |
Complex Structure Of Endo-1,3-Fucanase (Fun174Sb) From Gh174 Family With Fucotetraose
Organism: Spartobacteria bacterium
Method: X-RAY DIFFRACTION Release Date: 2024-12-18 Classification: HYDROLASE |
 |
Organism: Streptomyces sp. kcb13f003
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2023-11-08 Classification: HYDROLASE |
 |
Organism: Anaerolineae bacterium
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-10-19 Classification: PLANT PROTEIN Ligands: MPD, PO4, TRS |
 |
Solution Nmr Structure Of A Mutant Major Ampullate Spidroin 1 N-Terminal Domain
Organism: Euprosthenops australis
Method: SOLUTION NMR Release Date: 2019-09-11 Classification: STRUCTURAL PROTEIN |
 |
Crystal Structure Of An Asymmetric Dimer Of The N-Terminal Domain Of Euprosthenops Australis Major Ampullate Spidroin 1 (Dragline Silk)
Organism: Euprosthenops australis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-07-17 Classification: PROTEIN FIBRIL Ligands: SO4 |
 |
Crystal Structure Of A Gh2 Beta-Galacturonidase From Eisenbergiella Tayi Bound To Glycerol
Organism: Eisenbergiella tayi
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-02-20 Classification: HYDROLASE Ligands: CL, GOL |
 |
Crystal Structure Of Gh2 Beta-Galacturonidase From Eisenbergiella Tayi Bound To Galacturonate
Organism: Eisenbergiella tayi
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2019-02-20 Classification: HYDROLASE Ligands: ADA, CL |
 |
Bacterial Adhesin From Mobiluncus Mulieris Containing Intramolecular Disulfide, Isopeptide, And Ester Bond Cross-Links (Space Group P1)
Organism: Mobiluncus mulieris
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2018-06-13 Classification: CELL ADHESION |
 |
Bacterial Adhesin From Mobiluncus Mulieris Containing Intramolecular Disulfide, Isopeptide, And Ester Bond Cross-Links (Space Group P21)
Organism: Mobiluncus mulieris
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-06-13 Classification: CELL ADHESION |
 |
Organism: Euprosthenops australis
Method: SOLUTION NMR Release Date: 2013-11-27 Classification: STRUCTURAL PROTEIN |
 |
Structure Of Monomeric Nt From Euprosthenops Australis Major Ampullate Spidroin 1 (Masp1)
Organism: Euprosthenops australis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-08-01 Classification: STRUCTURAL PROTEIN Ligands: BR |
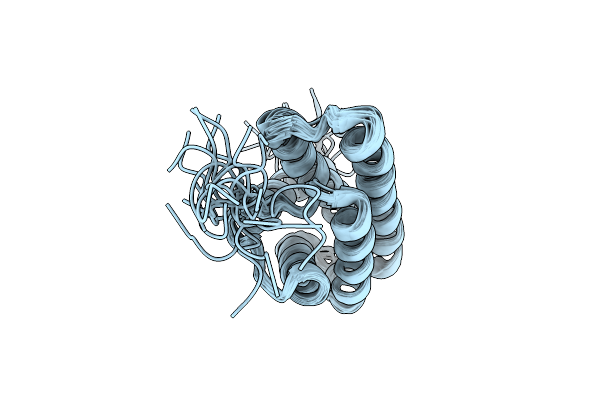 |
Nmr Structure Of A Monomeric Mutant (A72R) Of Major Ampullate Spidroin 1 N-Terminal Domain
Organism: Euprosthenops australis
Method: SOLUTION NMR Release Date: 2012-06-27 Classification: STRUCTURAL PROTEIN |
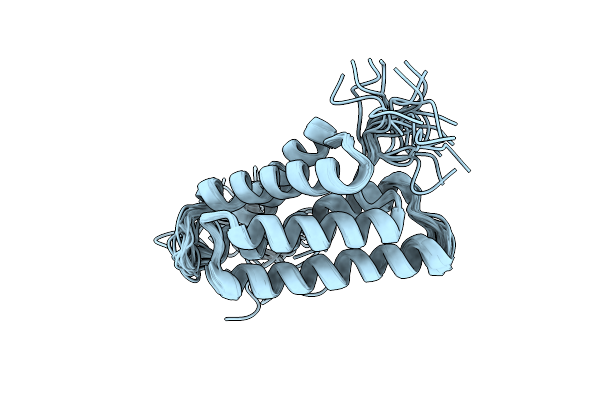 |
Organism: Euprosthenops australis
Method: SOLUTION NMR Release Date: 2012-06-27 Classification: STRUCTURAL PROTEIN |
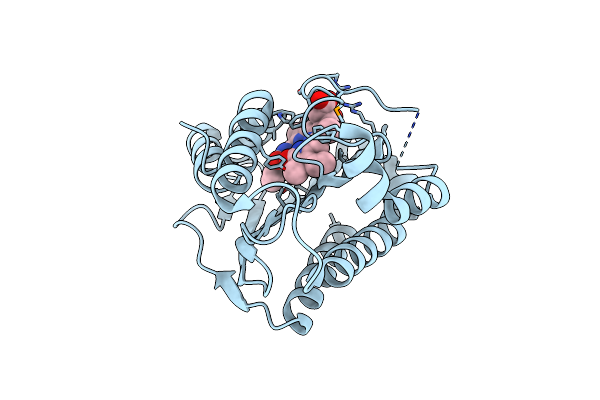 |
Organism: Synechococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2011-03-30 Classification: OXIDOREDUCTASE Ligands: BLA |
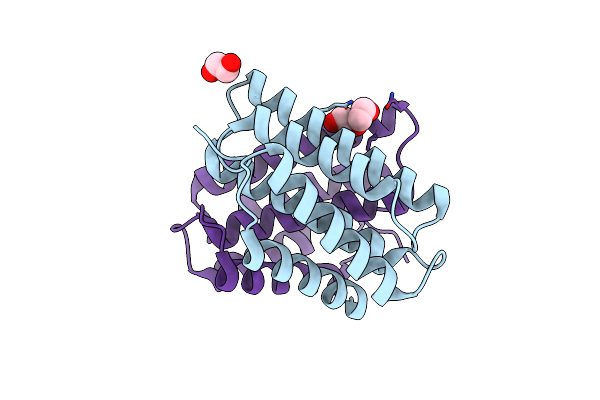 |
Self-Assembly Of Spider Silk Proteins Is Controlled By A Ph-Sensitive Relay
Organism: Euprosthenops australis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2010-05-12 Classification: STRUCTURAL PROTEIN Ligands: EDO, PEG |
 |
Self-Assembly Of Spider Silk Proteins Is Controlled By A Ph-Sensitive Relay
Organism: Euprosthenops australis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-05-12 Classification: STRUCTURAL PROTEIN Ligands: PGE |
 |
Self-Assembly Of Spider Silk Proteins Is Controlled By A Ph-Sensitive Relay
Organism: Euprosthenops australis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-05-12 Classification: STRUCTURAL PROTEIN Ligands: EDO, PEG, PGE |
 |
Self-Assembly Of Spider Silk Proteins Is Controlled By A Ph-Sensitive Relay
Organism: Euprosthenops australis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2010-05-12 Classification: STRUCTURAL PROTEIN Ligands: EDO, PGE |

