Search Count: 292
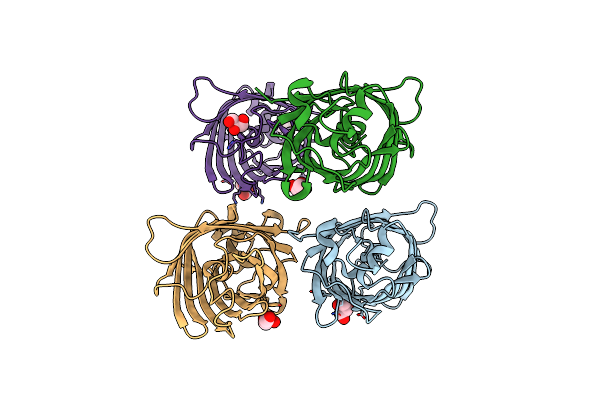 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2022-08-24 Classification: FLUORESCENT PROTEIN Ligands: GOL |
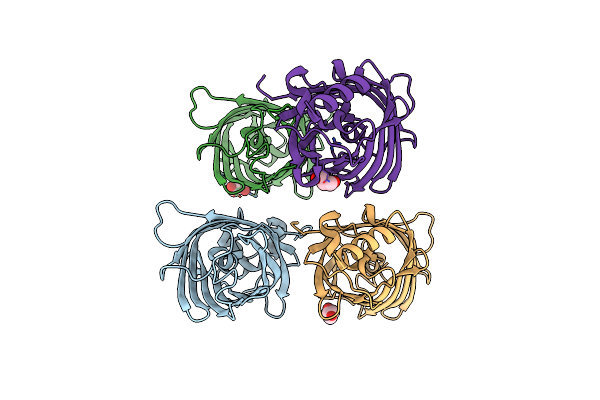 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2022-08-24 Classification: FLUORESCENT PROTEIN Ligands: GOL |
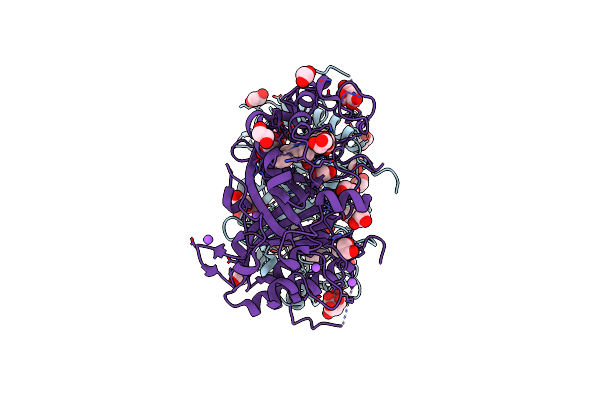 |
Crystal Structure Of Dye Decolorizing Peroxidase From Bacillus Subtilis At Acidic Ph
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-08-24 Classification: OXIDOREDUCTASE Ligands: HEM, OXY, EDO, CIT, PEG, NA, CL, GOL |
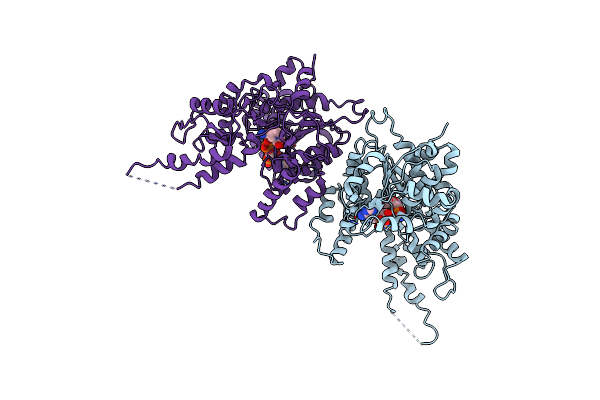 |
The Structure Of Rice Defective Pollen Wall (Dpw) In The Complex With Its Cofactor Nadp
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2022-08-24 Classification: OXIDOREDUCTASE Ligands: NAP |
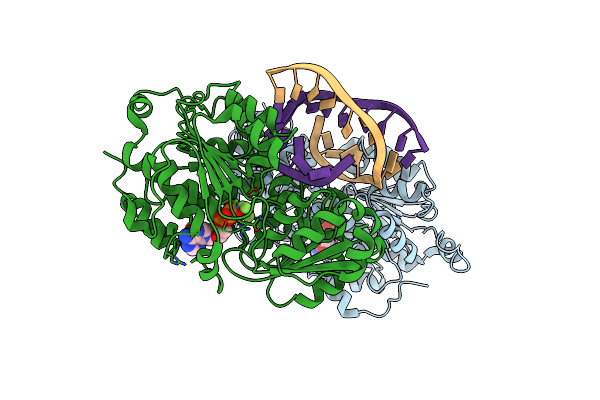 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-08-24 Classification: RNA-BINDING PROTEIN/DNA,RNA Ligands: MG, 08T |
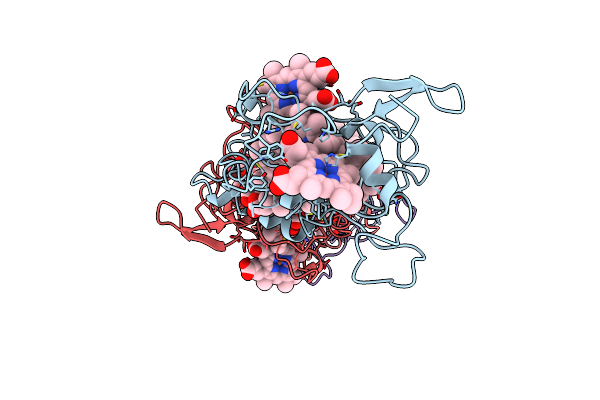 |
Organism: Geobacter sulfurreducens pca
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: PROTEIN FIBRIL Ligands: HEC |
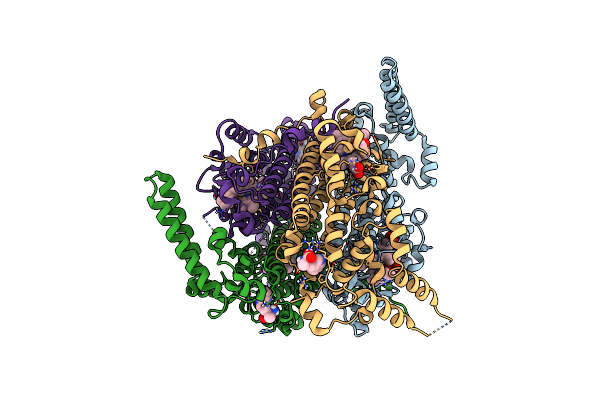 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-08-24 Classification: OXIDOREDUCTASE Ligands: HEM, ZIQ, 5PK |
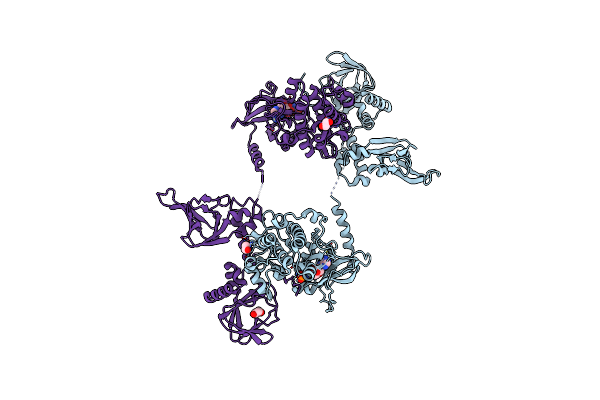 |
Crystal Structure Of Human Protein Kinase G (Pkg) R-C Complex In Inhibited State
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2022-08-24 Classification: TRANSFERASE/INHIBITOR Ligands: MN, ANP, EDO |
 |
Crystal Structure Of A Single-Chain E/F Type Bilin Lyase-Isomerase Mpeq In Space Group C2221
Organism: Synechococcus sp. a15-62
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2022-08-24 Classification: LYASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-08-24 Classification: HYDROLASE/INHIBITOR Ligands: MG, TLI |
 |
Organism: Synechococcus elongatus pcc 7942 = fachb-805
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2022-08-24 Classification: TRANSFERASE Ligands: IPE, EDO, PEG, SO4, MG, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-08-24 Classification: ANTIMICROBIAL PROTEIN Ligands: CL, CA, HEM, SCN, 8PR, PO4, NAG |
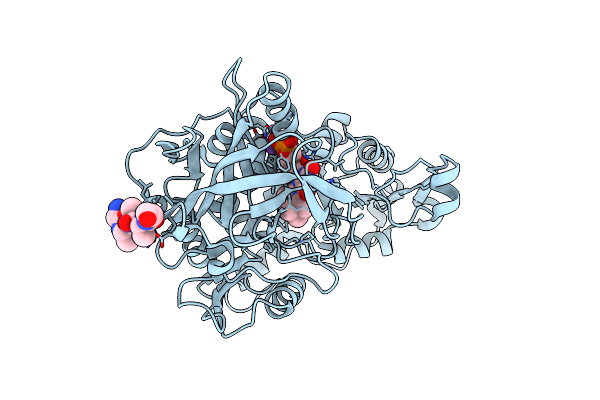 |
Crystal Structure Of Phenazine 1-Carboxylic Acid Decarboxylase From Mycobacterium Fortuitum
Organism: Mycolicibacterium fortuitum, Unidentified
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2022-08-24 Classification: LYASE Ligands: MN, 4LU, NA |
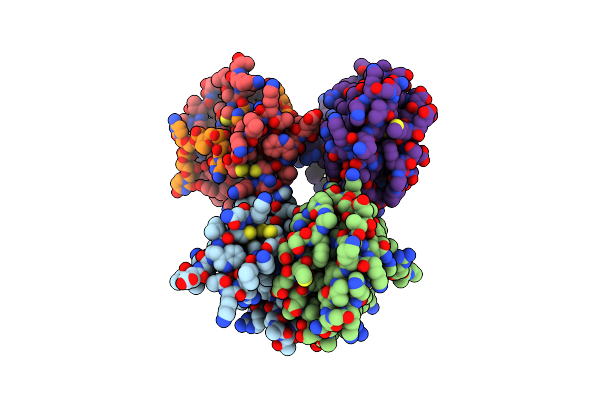 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2022-08-24 Classification: RNA BINDING PROTEIN Ligands: ZN |
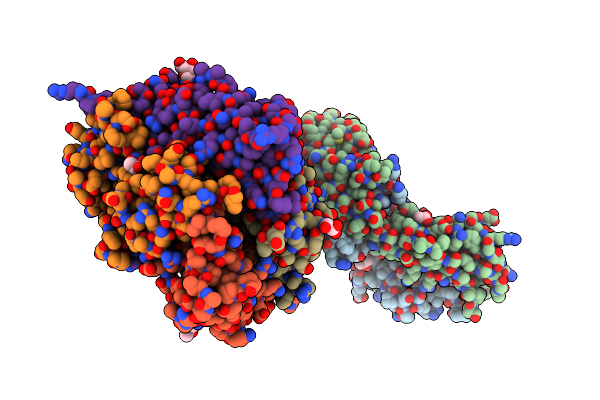 |
A Viral Peptide From Marek'S Disease Virus Bound To Chicken Mhc-Ii Molecule
Organism: Gallus gallus, Marek's disease herpesvirus (strain md11/75c/r2)
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2022-08-24 Classification: IMMUNE SYSTEM Ligands: NAG, GOL, ACT, PEG |
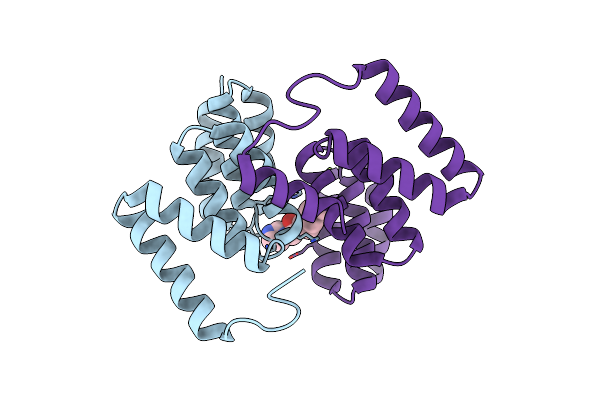 |
Organism: Shigella flexneri
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-08-24 Classification: CHAPERONE Ligands: CL, 483, MG |
 |
Organism: Shigella flexneri
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2022-08-24 Classification: CHAPERONE Ligands: CL, DMS, MG |
 |
Structure Of The Sporulation/Germination Protein Yhcn From Bacillus Subtilis
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2022-08-24 Classification: UNKNOWN FUNCTION |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2022-08-24 Classification: HYDROLASE Ligands: 1BO |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-08-24 Classification: HYDROLASE Ligands: GOL, SO4 |

