Search Count: 72
 |
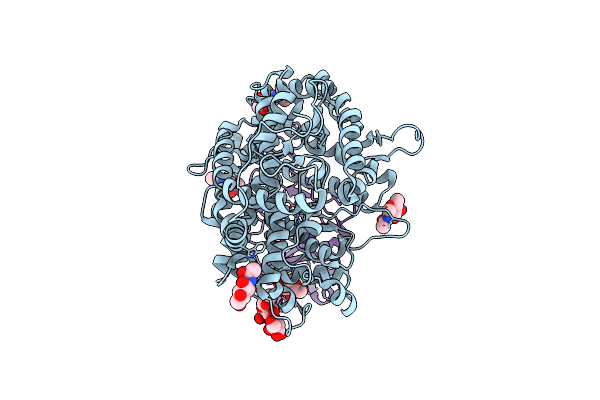
Cryo-Em Structure Of Hku25-Batcov S-Trimer Stabilized With 2P And X1 Disulfide Bond
Organism: Hypsugo bat coronavirus hku25, Tequatrovirus t4
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: VIRAL PROTEIN Ligands: EIC, NAG |
 |
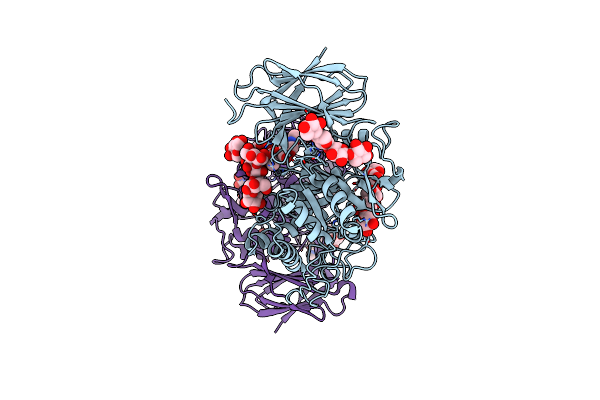
Organism: Bat sars-like virus btsy1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-23 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG, ZN |
 |
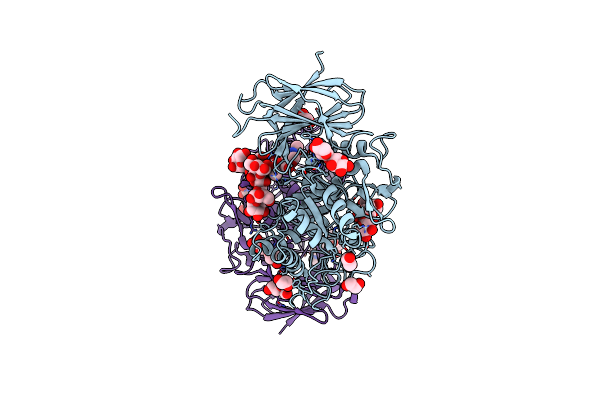
Crystal Structure Of Thermothelomyces Thermophila Gh30 (Double Mutant Ee) In Complex With Xylopentaose
Organism: Thermothelomyces
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-05-22 Classification: HYDROLASE Ligands: EDO, F |
 |
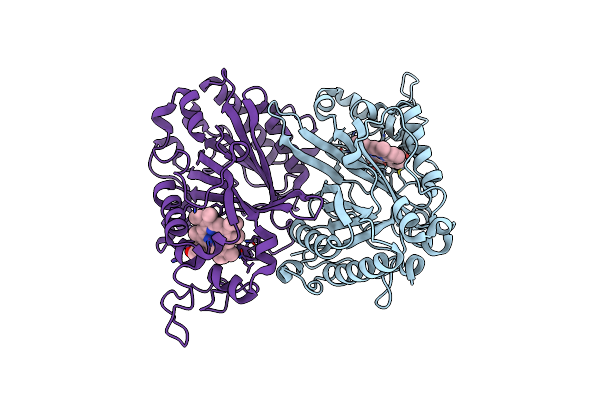
Crystal Structure Of Thermothelomyces Thermophila Gh30 (Double Mutant Ee) In Complex With Xylotriose.
Organism: Thermothelomyces
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-05-22 Classification: HYDROLASE Ligands: EDO, PEG |
 |
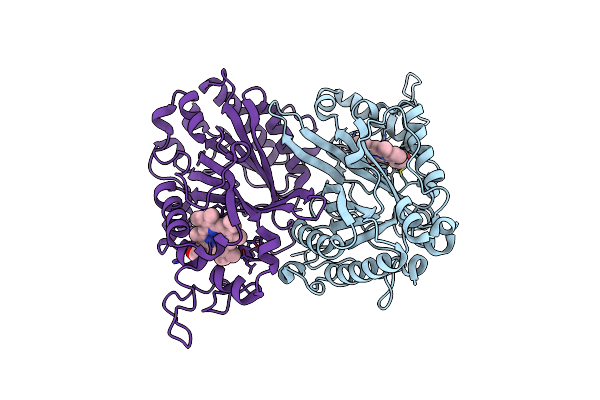
Structure Of Thermomonospora Curvata Heme-Containing Dyp-Type Peroxidase E293G Mutant
Organism: Thermomonospora curvata
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2023-11-29 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
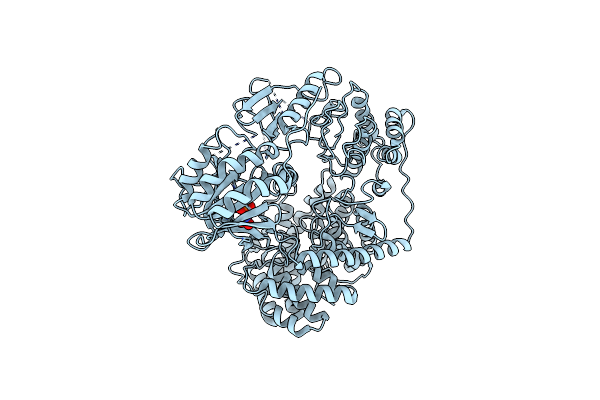
Structure Of Thermomonospora Curvata Heme-Containing Dyp-Type Peroxidase E293H Mutant
Organism: Thermomonospora curvata
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2023-11-29 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
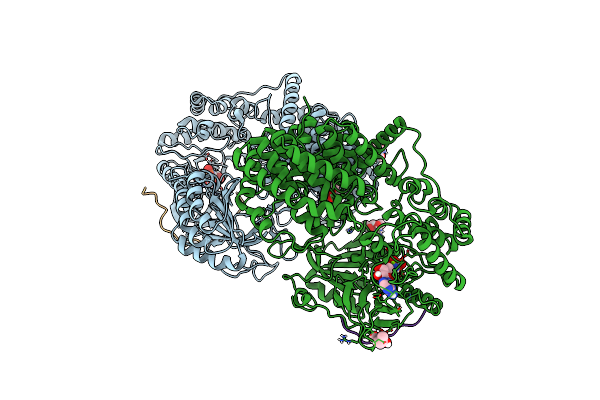
Discovery Of The Lanthipeptide Curvocidin And Structural Insights Into Its Trifunctional Synthetase Cuvl
Organism: Thermomonospora curvata
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2023-06-14 Classification: BIOSYNTHETIC PROTEIN Ligands: PO4, NO3 |
 |
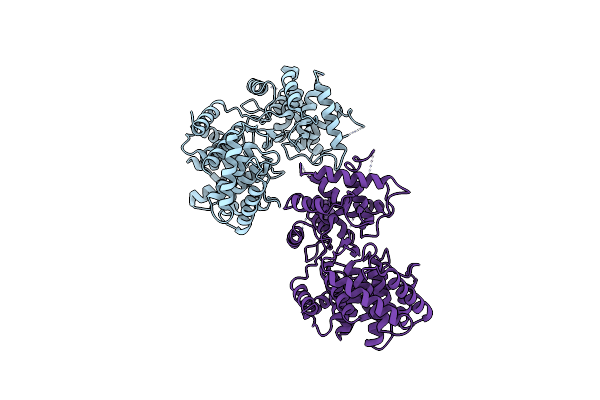
Discovery Of The Lanthipeptide Curvocidin And Structural Insights Into Its Trifunctional Synthetase Cuvl
Organism: Thermomonospora curvata
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2023-06-14 Classification: BIOSYNTHETIC PROTEIN Ligands: MPD, ANP, MG |
 |
Organism: Kitasatospora sp. cb02891
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2023-03-01 Classification: BIOSYNTHETIC PROTEIN |
 |
Structure Of Thermomonospora Curvata Heme-Containing Dyp-Type Peroxidase With A Modified Axial Ligand
Organism: Thermomonospora curvata
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-08-31 Classification: OXIDOREDUCTASE Ligands: HEM, SO4 |
 |
Crystal Structure Of Serine Hydroxymethyltransferase From Aphanothece Halophytica In The Covalent Complex With Malonate
Organism: Aphanothece halophytica
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2020-06-03 Classification: TRANSFERASE Ligands: PMP, EDO, MLI, NA |
 |
Crystal Structure Of Serine Hydroxymethyltransferase From Aphanothece Halophytica In The Plp-Internal Aldimine State
Organism: Aphanothece halophytica
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2020-06-03 Classification: TRANSFERASE Ligands: GOL, PEG |
 |
Crystal Structure Of Serine Hydroxymethyltransferase From Aphanothece Halophytica In The Plp-Serine External Aldimine State
Organism: Aphanothece halophytica
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2020-06-03 Classification: TRANSFERASE Ligands: GOL, PLS, 1PE |
 |
Biophysical And Structural Characterization Of The Thermostable Wd40 Domain Of A Prokaryotic Protein, Thermomonospora Curvata Pkwa
Organism: Thermomonospora curvata
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-10-03 Classification: PROTEIN BINDING |
 |
Structural Basis For The Catalytic Activity Of Thermomonospora Curvata Heme-Containing Dyp-Type Peroxidase.
Organism: Thermomonospora curvata
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2016-10-19 Classification: OXIDOREDUCTASE Ligands: HEM |
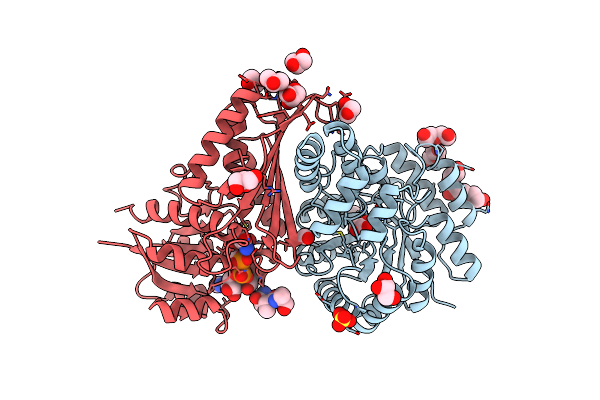 |
Crystal And Solution Structures Of The Bifunctional Enzyme (Aldolase/Aldehyde Dehydrogenase) From Thermomonospora Curvata, Reveal A Cofactor-Binding Domain Motion During Nad+ And Coa Accommodation Whithin The Shared Cofactor-Binding Site
Organism: Thermomonospora curvata
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2013-09-04 Classification: OXIDOREDUCTASE Ligands: SO4, PYR, CL, MG, GOL, PEG, NAD, FOR |
 |
 |
 |
 |

