Search Count: 145
 |
Organism: Ananas comosus
Method: SOLUTION NMR Release Date: 2011-10-26 Classification: HYDROLASE INHIBITOR |
 |
Organism: Homo sapiens, Mus musculus
Method: SOLUTION NMR Release Date: 2011-10-26 Classification: HYDROLASE, METAL BINDING PROTEIN Ligands: ZN |
 |
Organism: Mycobacterium tuberculosis
Method: SOLUTION NMR Release Date: 2011-10-26 Classification: OXIDOREDUCTASE |
 |
Solution Structure Of The Extracellular Domain Of The Tgf-Beta Type I Receptor
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2011-10-26 Classification: SIGNALING PROTEIN |
 |
Membrane Protein Complex Dsbb-Dsba Structure By Joint Calculations With Solid-State Nmr And X-Ray Experimental Data
Organism: Escherichia coli
Method: SOLID-STATE NMR Release Date: 2011-10-26 Classification: MEMBRANE PROTEIN, OXIDOREDUCTASE Ligands: ZN, UQ1 |
 |
Organism: Streptococcus sp. group g
Method: SOLID-STATE NMR Release Date: 2011-10-26 Classification: PROTEIN BINDING |
 |
Solution Nmr Structure Of Homeobox Domain (171-248) Of Human Homeobox Protein Tgif1, Northeast Structural Genomics Consortium Target Hr4411B
|
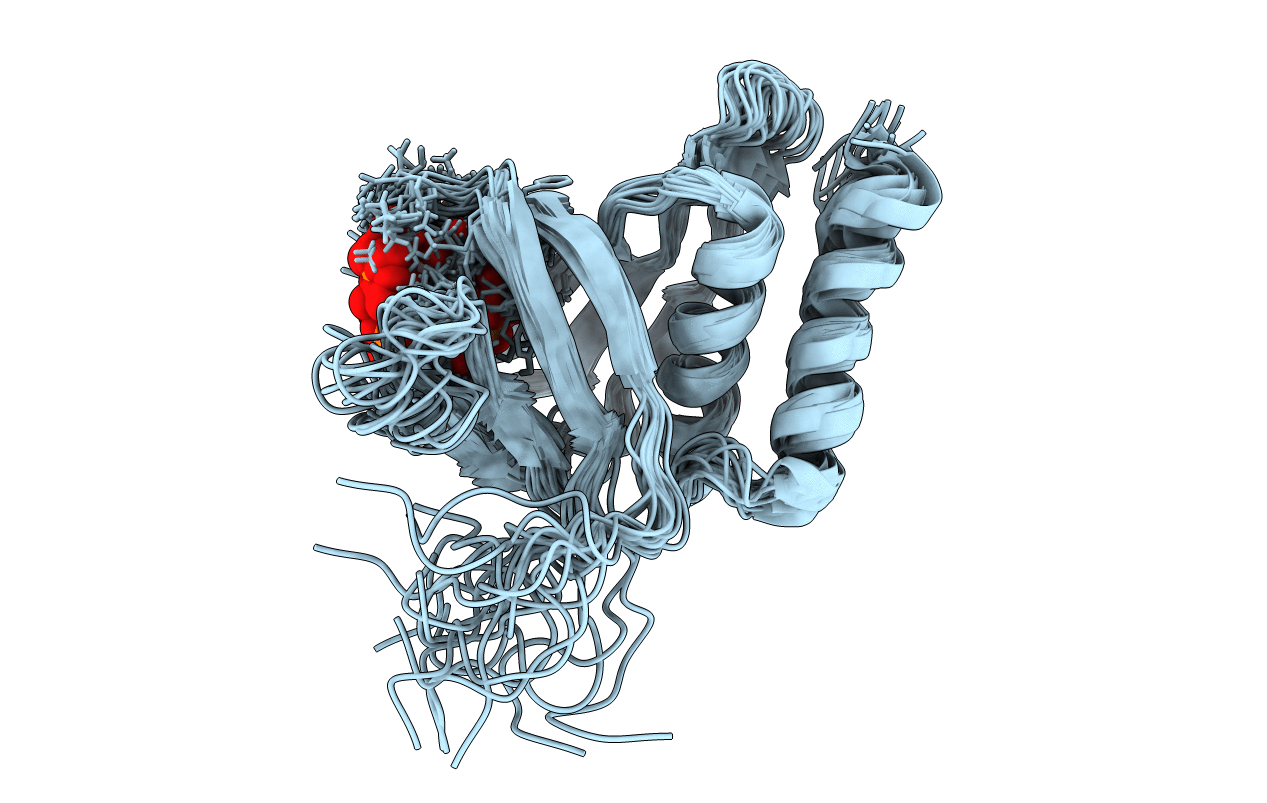 |
Structural Basis Of Phosphoinositide Binding To Kindlin-2 Pleckstrin Homology Domain In Regulating Integrin Activation
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2011-10-26 Classification: CELL ADHESION Ligands: 4IP |
 |
Organism: Aspergillus fumigatus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-10-26 Classification: HYDROLASE Ligands: AZM, PO4 |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2011-10-26 Classification: OXIDOREDUCTASE Ligands: CU, MPD, MRD, OH |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-10-26 Classification: OXIDOREDUCTASE Ligands: MPD, MRD |
 |
Crystal Structure Of Laccase From Thermus Thermophilus Hb27 Complexed With Hg, Crystal Of The Apoenzyme Soaked For 5 Min. In 5 Mm Hgcl2 At 278 K.
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-10-26 Classification: OXIDOREDUCTASE Ligands: HG, MPD, MRD |
 |
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-10-26 Classification: TRANSFERASE/VIRAL PROTEIN |
 |
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2011-10-26 Classification: TRANSFERASE/VIRAL PROTEIN |
 |
Structure Of Segment Klvffa From The Amyloid-Beta Peptide (Ab, Residues 16-21), Alternate Polymorph Iii
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-10-26 Classification: PROTEIN FIBRIL |
 |
Structure Of Segment Klvffa From The Amyloid-Beta Peptide (Ab, Residues 16-21), Alternate Polymorph I
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2011-10-26 Classification: PROTEIN FIBRIL Ligands: ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-10-26 Classification: APOPTOSIS Ligands: PEG, PGE |
 |
Solution Structure Of Aucg Tetraloop Hairpin Found In Human Xist Rna A-Repeats Essential For X-Inactivation
|
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-10-26 Classification: LIGASE/TRANSPORT PROTEIN Ligands: EDO |
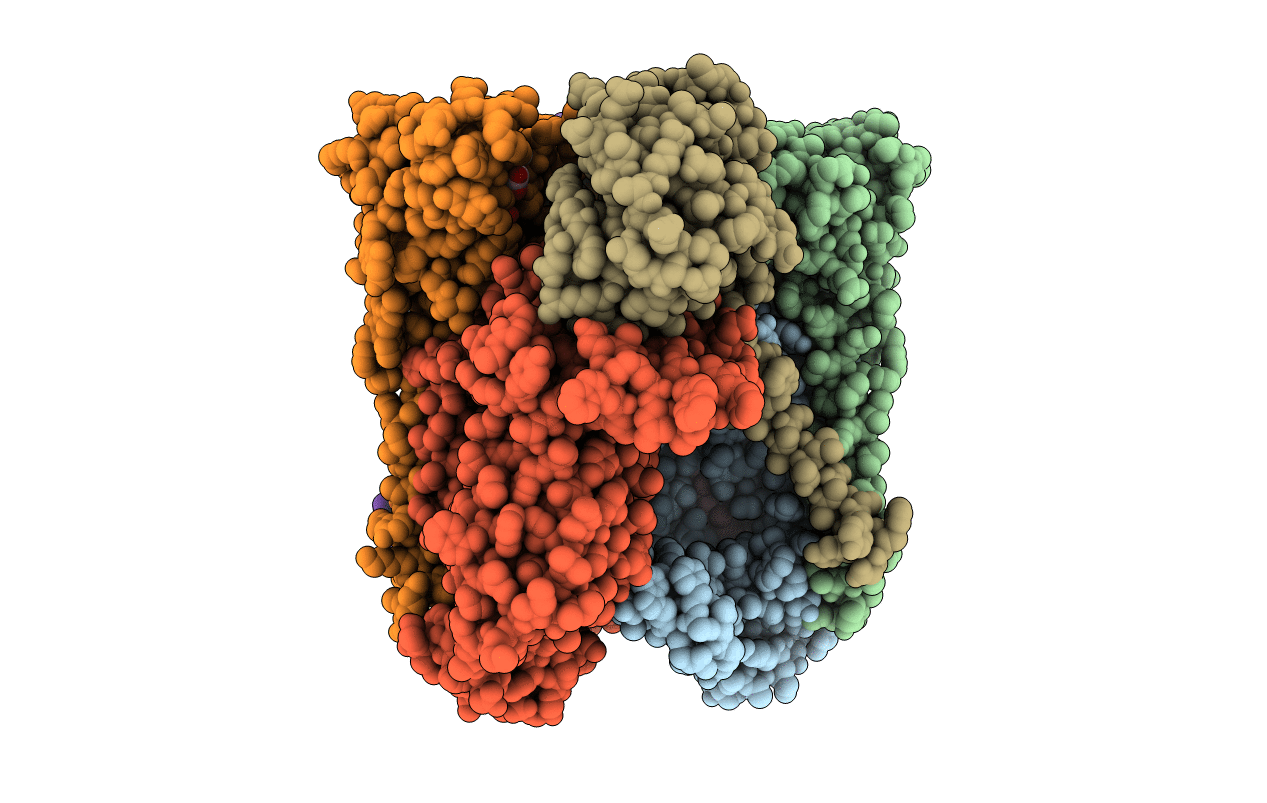 |
X-Ray Structure Of The Dimeric Cytochrome Bc1 Complex From The Soil Bacterium Paracoccus Denitrificans At 2.7 Angstrom Resolution
Organism: Paracoccus denitrificans
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2011-10-26 Classification: OXIDOREDUCTASE Ligands: HEM, SMA, HEC, FES |

