Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Sporormiella sp.
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: OXIDOREDUCTASE Ligands: FE, CL |
 |
The Ring Expansion Oxygenase Spoc In Complex With Mn And N-Oxalylglycine (Nog)
Organism: Sporormiella sp.
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: OXIDOREDUCTASE Ligands: MN, OGA, GOL |
 |
Organism: Sporormiella sp.
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: OXIDOREDUCTASE Ligands: FE, CL |
 |
Organism: Bacteroidetes bacterium hgw-bacteroidetes-12, Synthetic construct, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RNA BINDING PROTEIN Ligands: MG |
 |
Organism: Bacteroidetes bacterium hgw-bacteroidetes-12, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RNA BINDING PROTEIN |
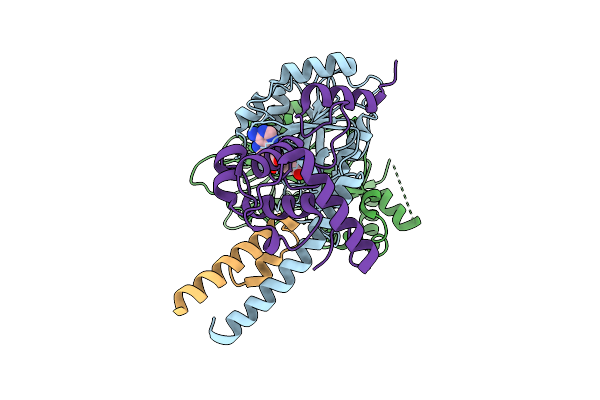 |
Organism: Tetrahymena thermophila sb210
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN Ligands: SAM |
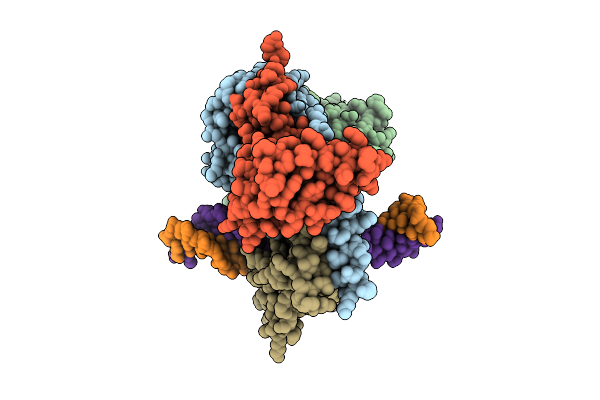 |
Cryo-Em Structure Of Hemimethylated Dna-Bound Tetrahymena Dna Methyltransferase Complex Mta1C
Organism: Tetrahymena thermophila sb210, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN/DNA Ligands: SAM |
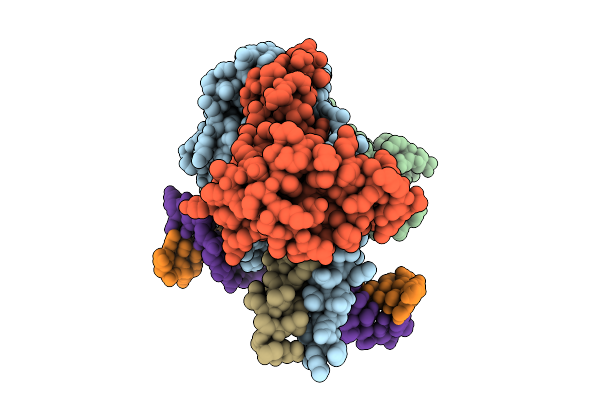 |
Cryo-Em Structure Of Hemi-Methylated Dna-Bound Tetrahymena Dna Methyltransferase Complex Mta1C (Mta9-B)
Organism: Tetrahymena thermophila sb210, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN/DNA Ligands: SAM |
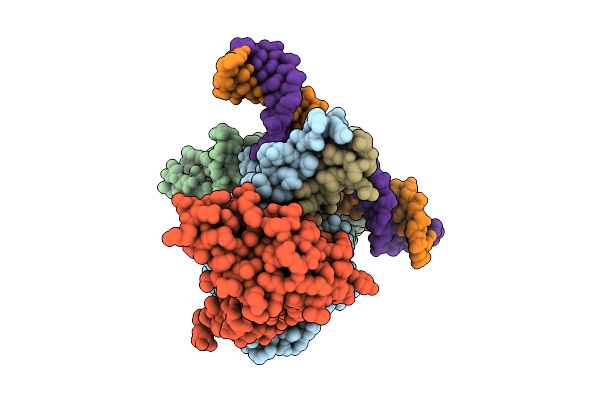 |
Cryo-Em Structure Of Unmethylated Dna-Bound Tetrahymena Dna Methyltransferase Complex Mta1C (Mta9-B)
Organism: Tetrahymena thermophila sb210, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN/DNA Ligands: SAM |
 |
Crystal Structure Of Ydjb From Salmonella Enterica Subsp. Enterica Serovar Typhi
Organism: Salmonella enterica subsp. enterica
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: DE NOVO PROTEIN |
 |
The Bifunctional Arabinofuranosidase/Xylosidase From Metagenome Of Pseudacanthotermes Militaris.
Organism: Pseudacanthotermes militaris
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: CA, TRS, GOL |
 |
Organism: Tetrahymena thermophila sb210
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: TRANSFERASE Ligands: SAH |
 |
Cryo-Em Structure Of The Zeaxanthin-Bound Light-Driven Proton Pumping Rhodopsin, Nm-R1
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, R16, D12, K3I |
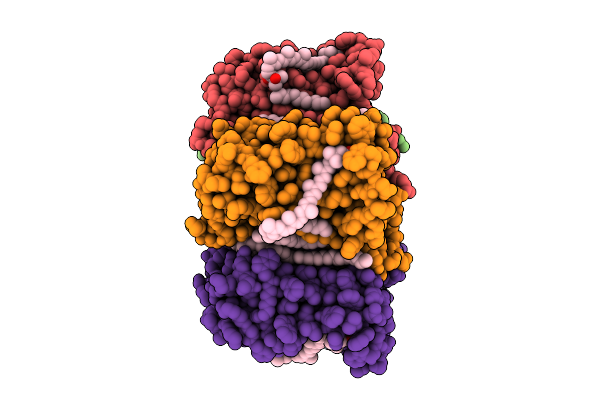 |
Cryo-Em Structure Of The Myxol-Bound Light-Driven Proton Pumping Rhodopsin, Nm-R1
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, R16, D12, A1L4O |
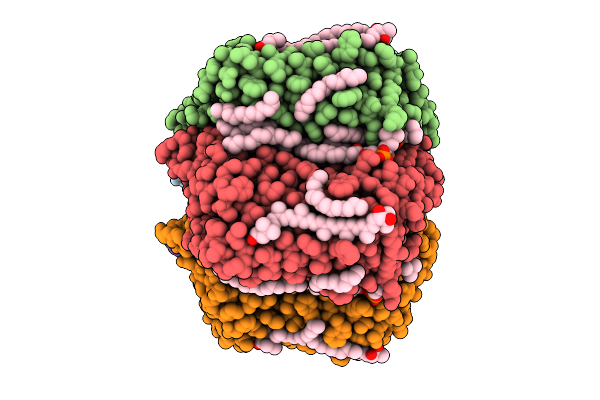 |
Cryo-Em Structure Of The Myxol-Bound Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, A1L4O, CL, PC1, PLC, R16, 8K6, D12, C14, D10 |
 |
Cryo-Em Structure Of The Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, CL, PC1, PLC, D12, R16, 8K6, C14 |
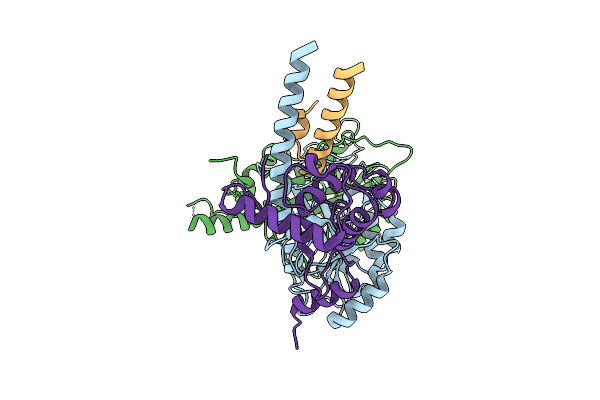 |
Organism: Tetrahymena thermophila sb210
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: DNA BINDING PROTEIN |
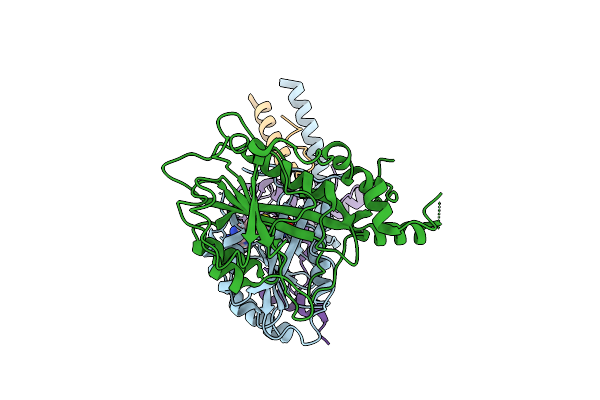 |
Cryo-Em Structure Of Sah-Bound Tetrahymena Dna Methyltransferase Complex Mta1C
Organism: Tetrahymena thermophila sb210
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: DNA BINDING PROTEIN Ligands: SAH |
 |
Cryo-Em Structure Of Sam-Bound Tetrahymena Dna Methyltransferase Complex Mta1C
Organism: Tetrahymena thermophila sb210
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: DNA BINDING PROTEIN Ligands: SAM |
 |
Cryo-Em Structure Of Sam-Bound Tetrahymena Dna Methyltransferase Complex Mta1C (D209A)
Organism: Tetrahymena thermophila sb210
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: DNA BINDING PROTEIN Ligands: SAM |