Search Count: 84
 |
Crystal Structure Of Sh3-Like_Bac-Type Domain (79-145) Of Conserved Domain Protein Gbaa_2967 From Bacillus Anthracis Ames Ancestor
Organism: Bacillus anthracis str. ames
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: UNKNOWN FUNCTION Ligands: MG, FMT |
 |
Organism: Bacillus anthracis str. ames
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2024-03-06 Classification: TRANSCRIPTION |
 |
2.9 Angstrom Resolution Crystal Structure Of Dtdp-Glucose 4,6-Dehydratase (Rfbb) From Bacillus Anthracis Str. Ames In Complex With Nad.
Organism: Bacillus anthracis str. ames
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2017-11-08 Classification: LYASE Ligands: NI, NAD, SO4 |
 |
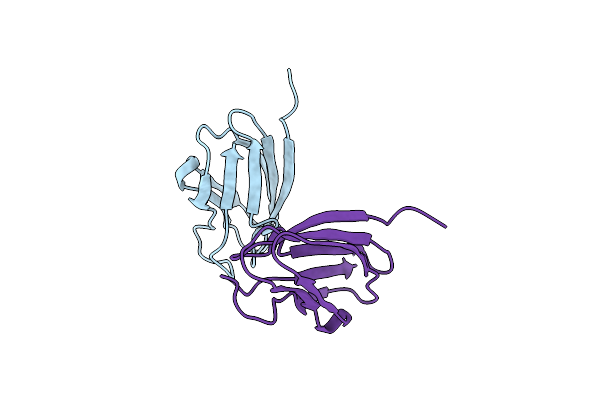
Crystal Structure Of Uncharacterized Cupredoxin-Like Domain Protein From Bacillus Anthracis
Organism: Bacillus anthracis str. ames
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2016-11-16 Classification: OXIDOREDUCTASE Ligands: CD, MLT, EDO, FMT |
 |
Crystal Structure Of Uncharacterized Cupredoxin-Like Domain Protein From Bacillus Anthracis
Organism: Bacillus anthracis str. ames
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2016-11-16 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Mccf-Like Protein (Ba_5613) In Complex With Asa (Alanyl Sulfamoyl Adenylates)
Organism: Bacillus anthracis str. ames
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2015-12-30 Classification: HYDROLASE Ligands: A5A |
 |
The Crystal Structure Of A Functionally Unknown Conserved Protein Mutant From Bacillus Anthracis Str. Ames
Organism: Bacillus anthracis str. ames
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2015-12-09 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Putative Glycerophosphoryl Diester Phosphodiesterasefrom Bacillus Anthraci
Organism: Bacillus anthracis str. ames
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2014-09-10 Classification: HYDROLASE Ligands: MG, SO4, EDO, GOL, PEG |
 |
Crystal Structure Of The Inosine 5'-Monophosphate Dehydrogenase, With A Internal Deletion Of Cbs Domain From Bacillus Anthracis Str. Ame Complexed With P32
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2014-07-23 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: IMP, 2F0, FMT, GOL, MLI, EDO, SO4 |
 |
Crystal Structure Of The Inosine 5'-Monophosphate Dehydrogenase With An Internal Deletion Of The Cbs Domain From Bacillus Anthracis Str. Ames Complexed With Inhibitor C91
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2014-06-11 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: IMP, C91, MLI |
 |
Crystal Structure Of Kynurenine Formamidase From Bacillus Anthracis Complexed With 2-Aminoacetophenone.
Organism: Bacillus anthracis str. ames
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2014-04-30 Classification: HYDROLASE Ligands: ZN, VNJ, MG |
 |
Crystal Structure Of The Double Mutant (S112A, H303A) Of B.Anthracis Mycrocine Immunity Protein (Mccf) With Aspartyl Sulfamoyl Adenylates
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2014-04-23 Classification: IMMUNE SYSTEM Ligands: DSZ, SO4 |
 |
The Crystal Structure Of A Solute Binding Protein From Bacillus Anthracis Str. Ames In Complex With Quorum-Sensing Signal Autoinducer-2 (Ai-2)
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2014-04-09 Classification: SUGAR BINDING PROTEIN Ligands: PAV, EDO, CL |
 |
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2014-04-02 Classification: HYDROLASE Ligands: ZN, MG, DIO, GXT |
 |
Crystal Structure Of The Inosine 5'-Monophosphate Dehydrogenase, With A Internal Deletion Of Cbs Domain From Bacillus Anthracis Str. Ames Complexed With P68
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-01-01 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: IMP, P68 |
 |
Crystal Structure Of The Inosine 5'-Monophosphate Dehydrogenase With An Internal Deletion Of The Cbs Domain From Bacillus Anthracis Str. Ames Complexed With Inhibitor A110
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-01-01 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: IMP, 2EY, GOL |
 |
1.8 Angstrom Crystal Structure Of Signal Peptidase I From Bacillus Anthracis.
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-12-18 Classification: HYDROLASE Ligands: PG4, PEG, EDO |
 |
Crystal Structure Of The Inosine 5'-Monophosphate Dehydrogenase With An Internal Deletion Of The Cbs Domain From Bacillus Anthracis Str. Ames Complexed With Inhibitor Q21
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2013-11-13 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: IMP, Q21, EDO, FMT, ACY |
 |
Crystal Structure Of The Inosine 5'-Monophosphate Dehydrogenase, With A Short Internal Deletion Of Cbs Domain From Bacillus Anthracis Str. Ames
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2013-10-09 Classification: OXIDOREDUCTASE Ligands: EDO |
 |
Crystal Structure Of The Double Mutant (S112A, H303A) Of B.Anthracis Mycrocine Immunity Protein (Mccf)
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2013-10-02 Classification: IMMUNE SYSTEM Ligands: GOL, EDO |

