Search Count: 657
 |
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: CELL CYCLE |
 |
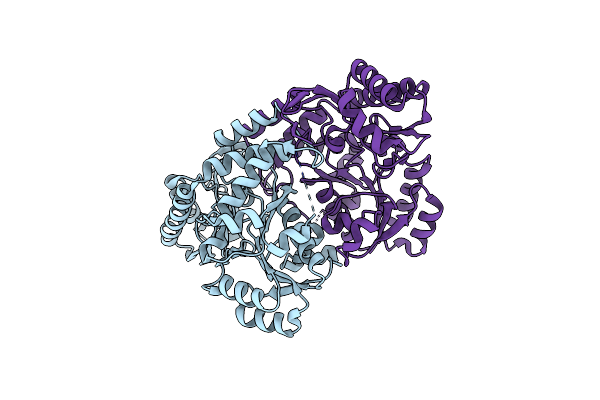
Corynebacterium Glutamicum Pyruvate:Quinone Oxidoreductase (Pqo), C-Terminal Truncated Construct
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2024-08-07 Classification: OXIDOREDUCTASE Ligands: FAD, TPP, MG |
 |
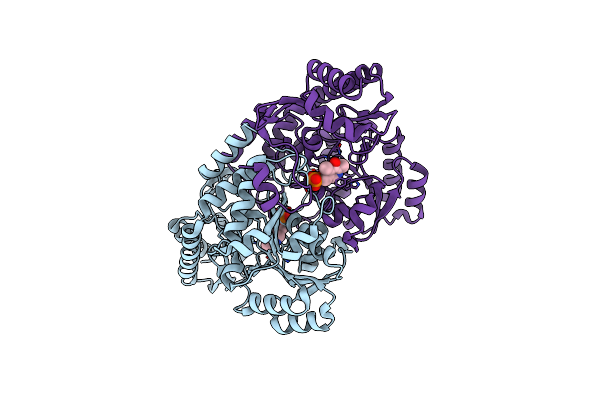
Crystal Structure Of Acetylornithine Aminotransferase From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-01-17 Classification: TRANSFERASE |
 |
Crystal Structure Of Acetylornithine Aminotransferase Complex With Plp From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2024-01-17 Classification: TRANSFERASE Ligands: PLP |
 |
Crystal Structure Of Apo-Form Of Dehydroquinate Dehydratase From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-12-27 Classification: LYASE Ligands: FLC, PG4 |
 |
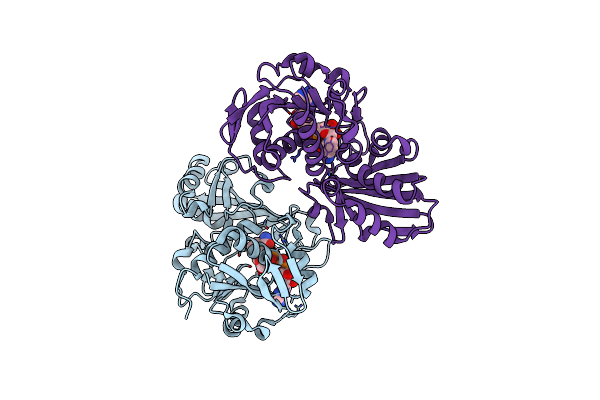
Crystal Structure Of Substrate Bound-Form Dehydroquinate Dehydratase From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-12-27 Classification: LYASE Ligands: SO4, DQA, GOL |
 |
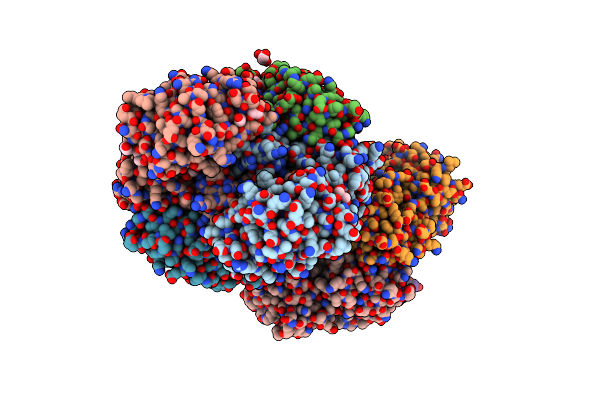
Crystal Structure Of Glyceraldehyde-3-Phosphate Dehydrogenase From Corynebacterium Glutamicum Atcc13032 In Complex With Nad
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2023-12-06 Classification: STRUCTURAL PROTEIN Ligands: NAD |
 |
Crystal Structure Of Glyceraldehyde-3-Phosphate Dehydrogenase From Corynebacterium Glutamicum Atcc13032 In Complex With Nad And G3P
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2023-12-06 Classification: STRUCTURAL PROTEIN Ligands: NAD, G3H, GOL, EDO, PG4 |
 |
Crystal Structure Of Glyceraldehyde-3-Phosphate Dehydrogenase From Corynebacterium Glutamicum Atcc13032 (L36S/T37K) In Complex With Nad
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2023-12-06 Classification: STRUCTURAL PROTEIN Ligands: CS, NAD, EDO, GOL, PO4 |
 |
Crystal Structure Of Glyceraldehyde-3-Phosphate Dehydrogenase From Corynebacterium Glutamicum Atcc13032 (L36S/T37K/F100V) In Complex With Nadp
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-12-06 Classification: STRUCTURAL PROTEIN Ligands: NAP, EDO |
 |
Crystal Structure Of Glyceraldehyde-3-Phosphate Dehydrogenase From Corynebacterium Glutamicum Atcc13032 (L36S/T37K/P192S) In Complex With Nadp
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-12-06 Classification: STRUCTURAL PROTEIN Ligands: NAP, GOL |
 |
Crystal Structure Of Glyceraldehyde-3-Phosphate Dehydrogenase From Corynebacterium Glutamicum Atcc13032 In Complex With Nadp
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2023-12-06 Classification: STRUCTURAL PROTEIN Ligands: NAP, GOL |
 |
C. Glutamicum S-Adenosylmethionine Synthase Co-Crystallized With Adenosine, Triphosphate, And Sam
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2023-10-25 Classification: TRANSFERASE Ligands: ADN, 3PO, SAM, GOL, POP, MG, K |
 |
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-10-25 Classification: TRANSFERASE Ligands: GOL, PEG, NA, PGE |
 |
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2023-10-25 Classification: TRANSFERASE Ligands: GOL, MES, NA, EDO |
 |
Crystal Structure Of The Homohexameric 2-Oxoglutarate Dehydrogenase Odha From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2023-08-16 Classification: OXIDOREDUCTASE Ligands: EPE, COA, ACO, TPP, MG |
 |
Single Particle Cryo-Em Structure Of The Homohexameric 2-Oxoglutarate Dehydrogenase Odha From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum atcc 13032
Method: ELECTRON MICROSCOPY Resolution:2.17 Å Release Date: 2023-08-16 Classification: OXIDOREDUCTASE Ligands: MG, ACO, TPP |
 |
Single Particle Cryo-Em Structure Of Homohexameric 2-Oxoglutarate Dehydrogenase Odha From Corynebacterium Glutamicum With Coenzyme A Bound To The E2O Domain
Organism: Corynebacterium glutamicum atcc 13032
Method: ELECTRON MICROSCOPY Resolution:2.17 Å Release Date: 2023-08-16 Classification: OXIDOREDUCTASE Ligands: MG, COA, ACO, TPP |
 |
Single Particle Cryo-Em Structure Of Homohexameric 2-Oxoglutarate Dehydrogenase Odha From Corynebacterium Glutamicum In Complex With The Product Succinyl-Coa
Organism: Corynebacterium glutamicum atcc 13032
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: OXIDOREDUCTASE Ligands: MG, ACO, TPP, SCA |
 |
Single Particle Cryo-Em Structure Of Homohexameric 2-Oxoglutarate Dehydrogenase Odha From Corynebacterium Glutamicum Following Reaction With The 2-Oxoglutarate Analogue Succinyl Phosphonate
Organism: Corynebacterium glutamicum atcc 13032
Method: ELECTRON MICROSCOPY Resolution:2.26 Å Release Date: 2023-08-16 Classification: OXIDOREDUCTASE Ligands: MG, ACO, QSP |

