Search Count: 945
 |
Organism: Clostridium perfringens atcc 13124
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: HYDROLASE Ligands: SIA, EDO |
 |
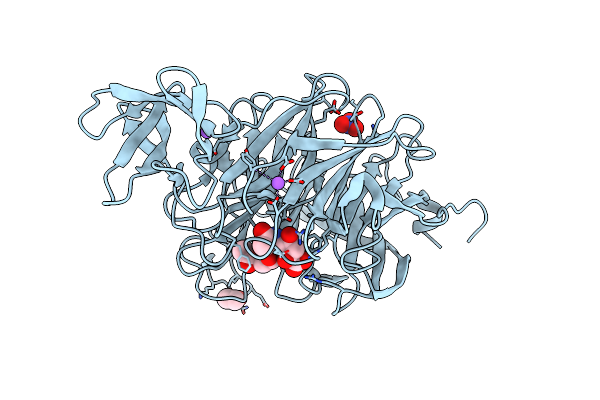
Crystal Structure Of Sugar Phosphotransferase System Eiib Component Cpf_0401 From Clostridium Perfringens
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-04-16 Classification: TRANSFERASE Ligands: TRS |
 |
Organism: Clostridium perfringens atcc 13124
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2024-09-04 Classification: HYDROLASE Ligands: NO3, NA, EDO, 5N6 |
 |
The Structure Of The Catalytic Domain Of Nani Sialdase In Complex With Neu5Gc
Organism: Clostridium perfringens atcc 13124
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-09-04 Classification: HYDROLASE Ligands: NGC, CA, NO3 |
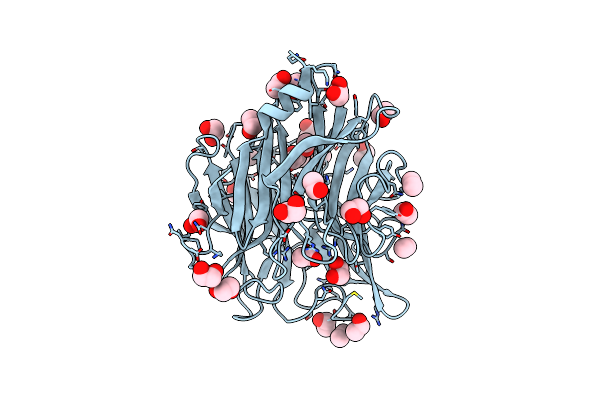 |
Organism: Clostridium perfringens atcc 13124
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-09-04 Classification: HYDROLASE Ligands: EDO, ACE |
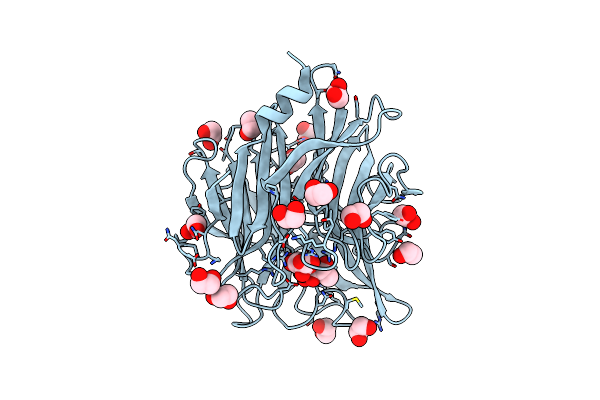 |
Organism: Clostridium perfringens atcc 13124
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-09-04 Classification: HYDROLASE Ligands: EDO, SIA |
 |
Organism: Clostridium perfringens atcc 13124
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2024-09-04 Classification: HYDROLASE Ligands: 5N6, EDO |
 |
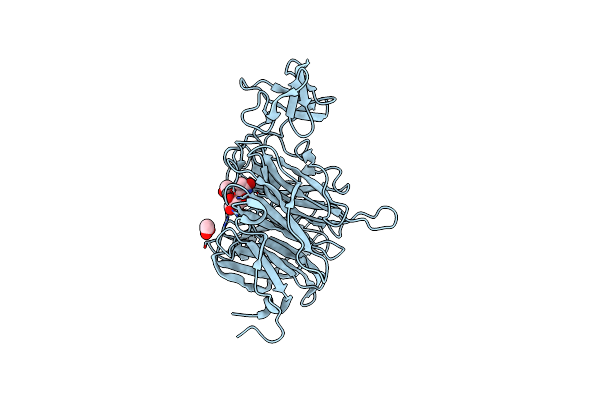
Organism: Clostridium perfringens atcc 13124
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2024-09-04 Classification: HYDROLASE Ligands: X1F, EDO |
 |
Organism: Clostridium perfringens atcc 13124
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-09-04 Classification: HYDROLASE Ligands: EDO, 5N6 |
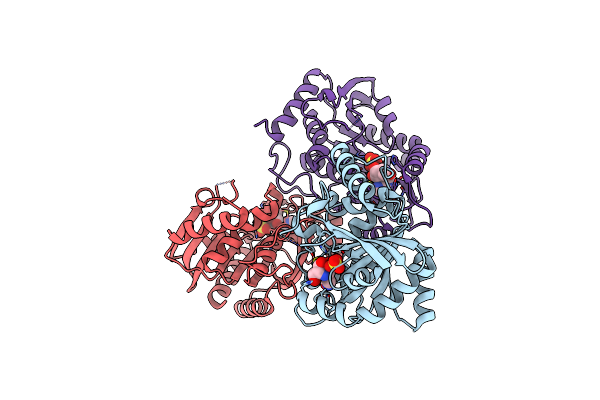 |
Structure Of Clostridium Perfringens Pnp Bound To Transition State Analog Immucillin H And Sulfate
Organism: Clostridium perfringens atcc 13124
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2023-10-18 Classification: TRANSFERASE Ligands: IMH, SO4 |
 |
X-Ray Structure Of Clostridium Perfringens Pili Protein B Collagen-Binding Domains
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2023-06-07 Classification: PROTEIN FIBRIL Ligands: ACT |
 |
Organism: Clostridium perfringens (strain 13 / type a)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-05-04 Classification: HYDROLASE Ligands: GLU, ZN, NA |
 |
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2021-07-28 Classification: HYDROLASE/INHIBITOR Ligands: EDO, CA, SO4, Z7S |
 |
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-07-28 Classification: HYDROLASE/INHIBITOR Ligands: EDO, CA, SO4, Z8V |
 |
The Structure Of The Catalytic Module Of The Metalloprotease Zmpa From Clostridium Perfringens
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-02-03 Classification: HYDROLASE Ligands: MES, EDO |
 |
The Structure Of The Cbm32-1, Cbm32-2, And M60 Catalytic Domains From Clostridium Perfringens Zmpb
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:4.60 Å Release Date: 2021-02-03 Classification: HYDROLASE Ligands: ZN |
 |
The Structure Of Cbm32-1 And Cbm32-2 Domains From Clostridium Perfringens Zmpb In Complex With Galnac
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: CA, A2G, GOL |
 |
The Structure Of Cbm32-1 And Cbm32-2 Domains From Clostridium Perfringens Zmpb
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: CA, GOL |
 |
The Structure Of Cbm32-1 And Cbm32-2 Domains From Clostridium Perfringens Zmpb In Complex With Galnac
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: A2G, CA, GOL |
 |
The Structure Of Cbm51-2 In Complex With Glcnac And Int Domains From Clostridium Perfringens Zmpb
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-02-03 Classification: SUGAR BINDING PROTEIN Ligands: CA, NAG, EDO, PO4, TRS |

