Search Count: 134
 |
Organism: Streptomyces rochei
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-12-04 Classification: CYTOSOLIC PROTEIN |
 |
Organism: Streptomyces rochei
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-12-04 Classification: BIOSYNTHETIC PROTEIN Ligands: ATP, MG, ACY, GOL, PO4 |
 |
Organism: Streptomyces rochei
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-12-04 Classification: BIOSYNTHETIC PROTEIN Ligands: ADP, PO4, MG, ACY, GOL, PEG, NA |
 |
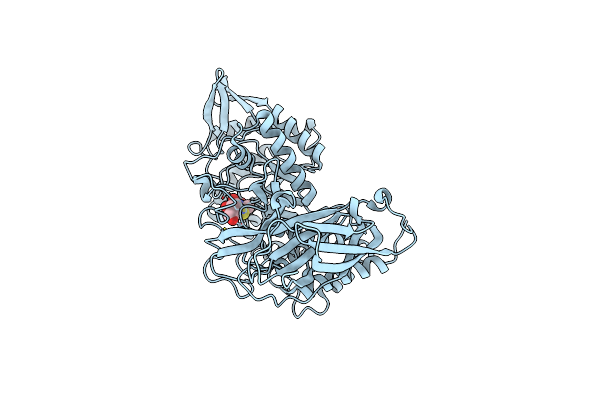
Crystal Structure Of Group 4 Monosaccharide-Releasing Beta-N-Acetylgalactosaminidase Ngap2 From Paenibacillus Sp. Ts12, Apo Form
Organism: Paenibacillus sp. ts12
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-04-24 Classification: HYDROLASE Ligands: BR |
 |
Crystal Structure Of Group 4 Monosaccharide-Releasing Beta-N-Acetylgalactosaminidase Ngap2 From Paenibacillus Sp. Ts12 In Complex With Galnac-Thiazoline
Organism: Paenibacillus sp. ts12
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-04-24 Classification: HYDROLASE Ligands: GNL, BR |
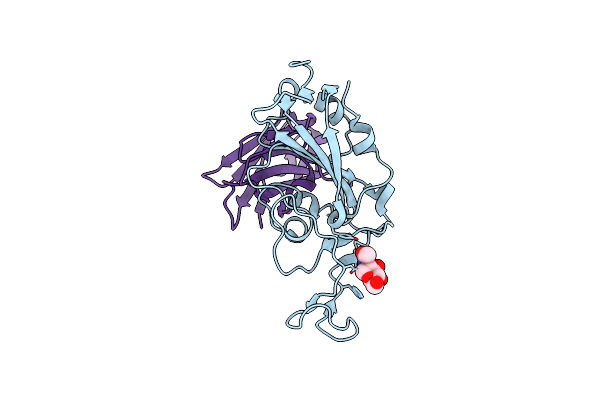 |
The Crystal Structure Of Bat Coronavirus Rsyn04 Rbd Bound To The Antibody S43
Organism: Betacoronavirus sp., Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-11-22 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
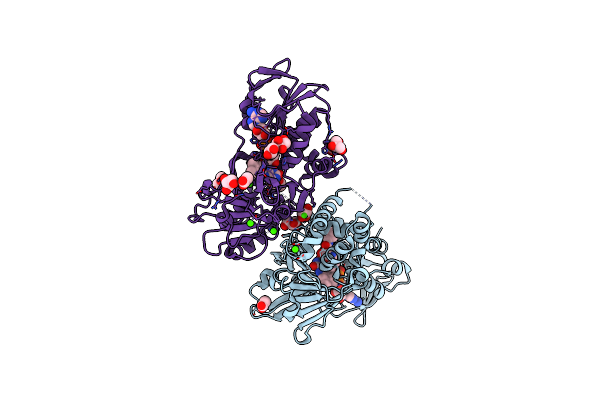 |
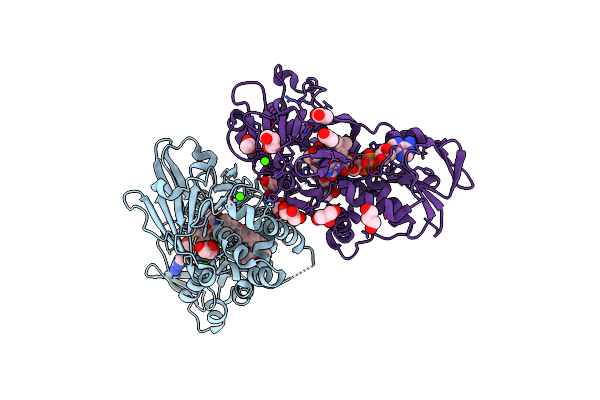
Crystal Structure Of A Dual Function Amine Oxidase/Cyclase In Complex With Substrate Analogues
Organism: Streptomyces rochei
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-09-19 Classification: FLAVOPROTEIN Ligands: FAD, ACT, CJ8, CA, GOL, CJE |
 |
Organism: Streptomyces rochei
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-09-19 Classification: FLAVOPROTEIN Ligands: FAD, CA, CWH, GOL, ACT |
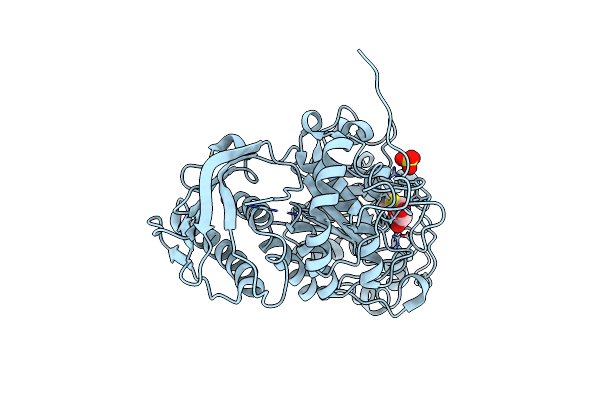 |
Crystal Structure Of Beta-Hexosaminidase From Paenibacillus Sp. Ts12 In Complex With Nag-Thiazoline.
Organism: Paenibacillus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-06-06 Classification: HYDROLASE Ligands: SO4, NGT |
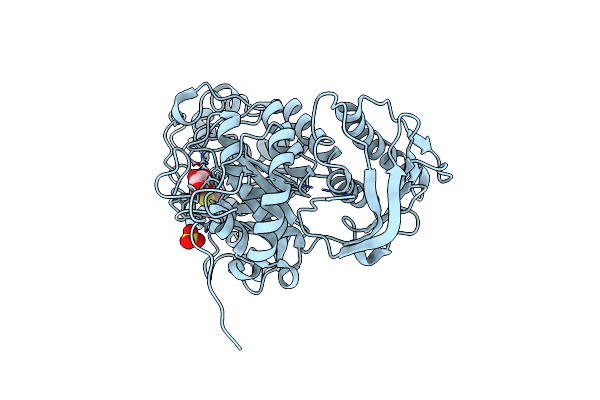 |
Crystal Structure Of Beta-Hexosaminidase From Paenibacillus Sp. Ts12 In Complex With Gal-Nag-Thiazoline
Organism: Paenibacillus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-06-06 Classification: HYDROLASE Ligands: SO4, GNL |
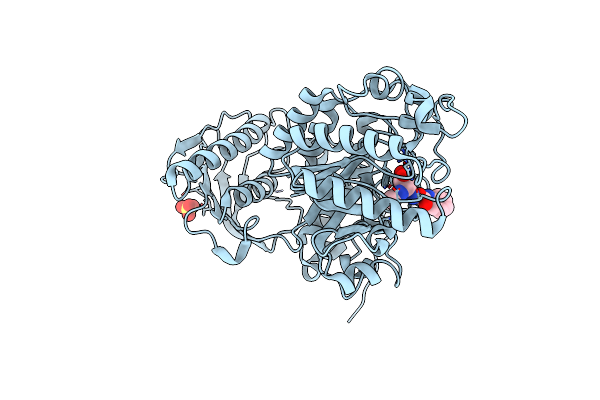 |
Crystal Structure Of Beta-Hexosaminidase From Paenibacillus Sp. Ts12 In Complex With Pugnac
Organism: Paenibacillus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-06-06 Classification: HYDROLASE Ligands: SO4, OAN |
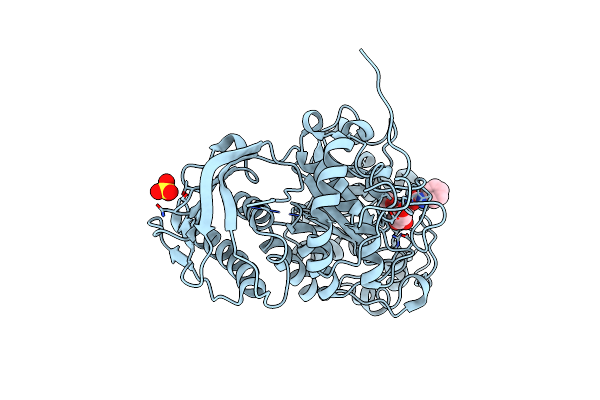 |
Crystal Structure Of Beta-Hexosaminidase From Paenibacillus Sp. Ts12 In Complex With Gal-Pugnac
Organism: Paenibacillus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2012-06-06 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: SO4, OGN |
 |
Crystal Structure Of Beta-Hexosaminidase From Paenibacillus Sp. Ts12 In Complex With Nhac-Dnj
Organism: Paenibacillus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2012-06-06 Classification: HYDROLASE Ligands: SO4, NOK |
 |
Crystal Structure Of Beta-Hexosaminidase From Paenibacillus Sp. Ts12 In Complex With Nhac-Cas
Organism: Paenibacillus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-06-06 Classification: HYDROLASE Ligands: SO4, GC2 |
 |
 |
 |
 |
 |
 |

