Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Shewanella putrefaciens cn-32
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE Ligands: MG |
 |
Organism: Shewanella putrefaciens cn-32
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2025-03-26 Classification: SIGNALING PROTEIN |
 |
Organism: Shewanella putrefaciens
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2024-03-20 Classification: SIGNALING PROTEIN Ligands: GDP |
 |
Dihydrodipicolinate Synthase (Dhdps) From C.Jejuni, Y110F Mutant With R,R-Bislysine Bound At The Allosteric Site At 2.7 Angstrom
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-03-16 Classification: LYASE Ligands: MG, PGE, 3VN, PEG, EDO |
 |
Glyceraldehyde 3-Phosphate Dehydrogenase From Campylobacter Jejeuni - Nad(P) Complex
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: NAP, PEG, NA |
 |
Glyceraldehyde 3-Phosphate Dehydrogenase From Campylobacter Jejeuni - Adp Complex
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: SO4, ADP, BME, ACT |
 |
Dihydrodipicolinate Synthase (Dhdps) From C.Jejuni, H59K Mutant With Pyruvate Bound In The Active Site In C2221 Space Group
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2022-01-12 Classification: LYASE Ligands: MG, EDO, PGE, ACT, PEG |
 |
Dihydrodipicolinate Synthase (Dhdps) From C.Jejuni, With Pyruvate Bound In The Active Site And L-Lysine Bound At The Allosteric Site In C121 Space Group
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2022-01-12 Classification: LYASE Ligands: LYS, MG |
 |
Dihydrodipicolinate Synthase (Dhdps) From C.Jejuni With Pyruvate Bound In The Active Site In P1211 Space Group
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2021-12-22 Classification: LYASE Ligands: PGE, MG, ACT, EDO |
 |
Dihydrodipicolinate Synthase (Dhdps) From C.Jejuni, Y110F Mutant With Pyruvate Bound In The Active Site And L-Lysine Bound At The Allosteric Site In C2221 Space Group
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-12-15 Classification: LYASE Ligands: LYS, PEG, EDO, ACT, PGE, MG |
 |
Dihydrodipicolinate Synthase (Dhdps) From C.Jejuni, H56N Mutant With Pyruvate Bound In The Active Site And R,R-Bislysine Bound At The Allosteric Site
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2021-12-08 Classification: LYASE Ligands: 3VN, ACT, PEG, PGE |
 |
Dihydrodipicolinate Synthase From C. Jejuni With Pyruvate Bound To The Active Site In C2221 Space Group
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2021-12-08 Classification: LYASE Ligands: PGE, GOL, ACT, EDO, PEG, MG |
 |
Dihydrodipicolinate Synthase (Dhdps) From C.Jejuni With Pyruvate Bound In The Active Site And L-Lysine Bound At The Allosteric Site In C2221 Space Group
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2021-12-08 Classification: LYASE Ligands: LYS, MG, EDO, PEG, PGE |
 |
Dihydrodipicolinate Synthase (Dhdps) From C.Jejuni, Y110F Mutant With Pyruvate Bound In The Active In C2221 Space Group
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2021-12-08 Classification: LYASE Ligands: PGE, MG, EDO, ACT, PEG |
 |
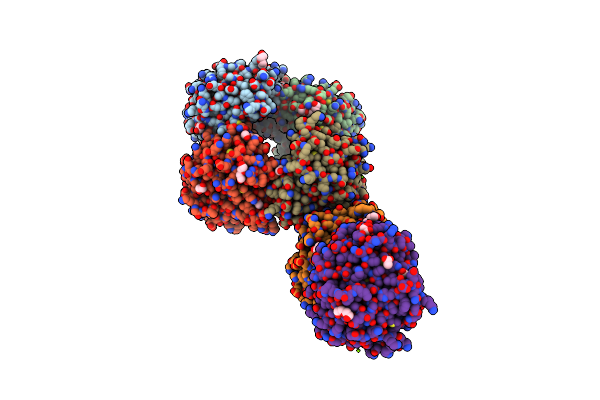
Dihydrodipicolinate Synthase (Dhdps) From C.Jejuni With Pyruvate Bound In The Active Site And L-Histidine Bound At The Allosteric Site
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2021-12-08 Classification: LYASE Ligands: MG, PGE, PEG, EDO, HIS, ACT, GOL |
 |
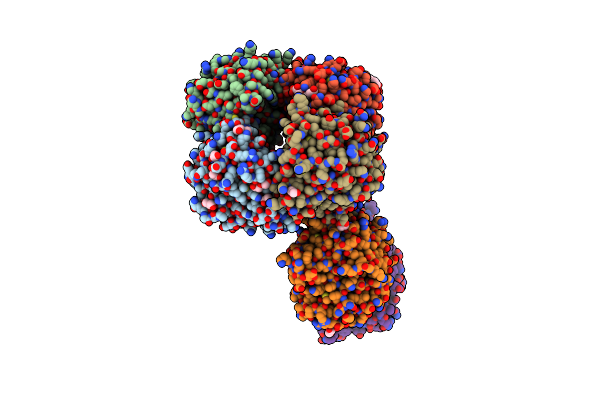
Dihydrodipicolinate Synthase (Dhdps) From C.Jejuni With Pyruvate Bound In The Active Site And R,R-Bislysine Bound At The Allosteric Site In C2221 Space Group
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2021-12-08 Classification: LYASE Ligands: 3VN, MG, PGE, ACT, PEG, EDO |
 |
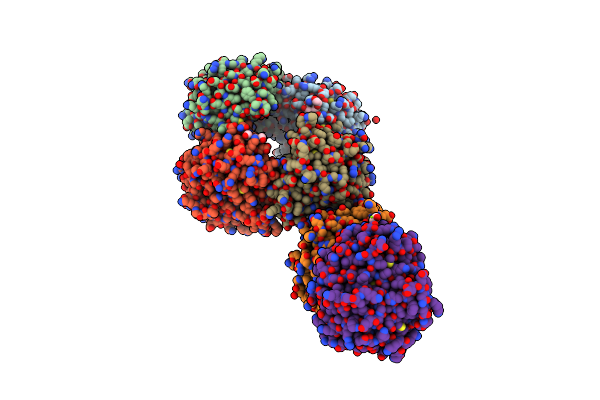
Dihydrodipicolinate Synthase (Dhdps) From C.Jejuni, H56A Mutant With Pyruvate Bound In The Active Site And R,R-Bislysine Bound At The Allosteric Site
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2021-12-01 Classification: LYASE Ligands: 3VN, EDO, PGE, GOL, PEG, ACT, MG |
 |
Dihydrodipicolinate Synthase (Dhdps) From C.Jejuni, H56N Mutant With Pyruvate Bound In The Active Site
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2021-12-01 Classification: LYASE Ligands: PGE, MG, EDO |
 |
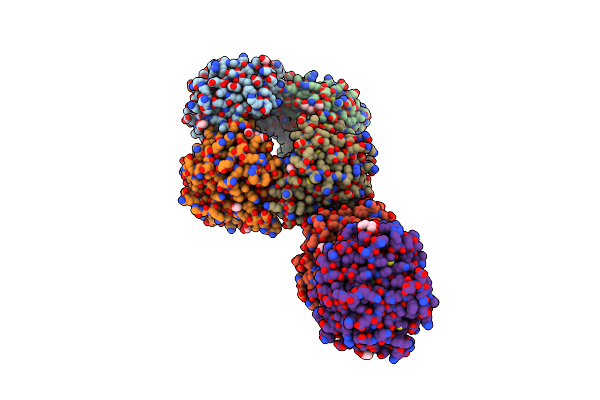
Dihydrodipicolinate Synthase (Dhdps) From C.Jejuni, H56N Mutant With Pyruvate Bound In The Active Site And L-Lysine Bound At The Allosteric Site
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2021-12-01 Classification: LYASE Ligands: LYS, PEG, EDO, ACT, MG, PGE, GOL |
 |
Dihydrodipicolinate Synthase (Dhdps) From C.Jejuni, H56A Mutant With Pyruvate Bound In The Active Site And L-Lysine At The Allosteric Site
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2021-11-24 Classification: LYASE Ligands: LYS, MG, PGE, EDO, ACT, PEG, GOL |