Search Count: 282
 |
Organism: Streptomyces lincolnensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-08-14 Classification: LYASE Ligands: GOL |
 |
Crystal Structure Of 3-Ketosteroid Delta1-Dehydrogenase From Rhodococcus Erythropolis Sq1 In Complex With 1,4-Androstadiene-3,17- Dione
Organism: Rhodococcus electrodiphilus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-06-26 Classification: OXIDOREDUCTASE Ligands: FAD, ANB |
 |
Organism: Streptomyces lincolnensis
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2024-04-03 Classification: TRANSFERASE |
 |
Structure Of Glycosyltransferase Lmbt In Complex With Gdp And Ergothioneine
Organism: Streptomyces lincolnensis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-04-03 Classification: TRANSFERASE Ligands: LW8, GDP |
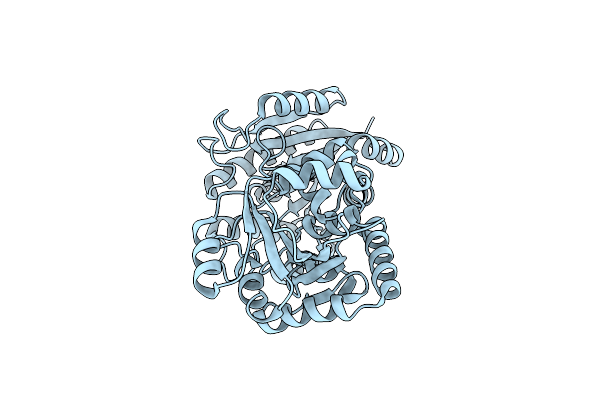 |
Organism: Streptomyces lincolnensis
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2023-09-20 Classification: BIOSYNTHETIC PROTEIN |
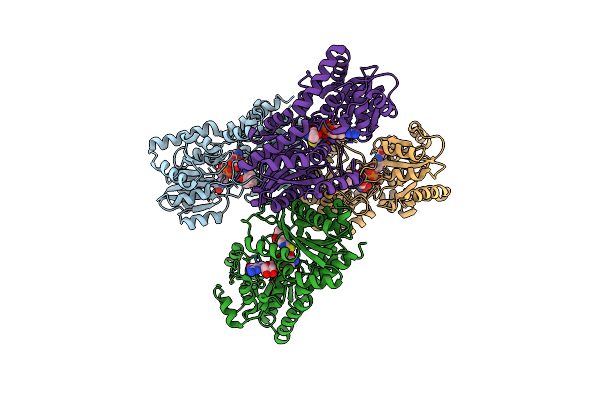 |
Crystal Structure Of Lmbt From Streptomyces Lincolnensis Nrrl Isp-5355 In Complex With Substrates
Organism: Streptomyces lincolnensis
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2023-09-20 Classification: BIOSYNTHETIC PROTEIN Ligands: GDP, Q3L |
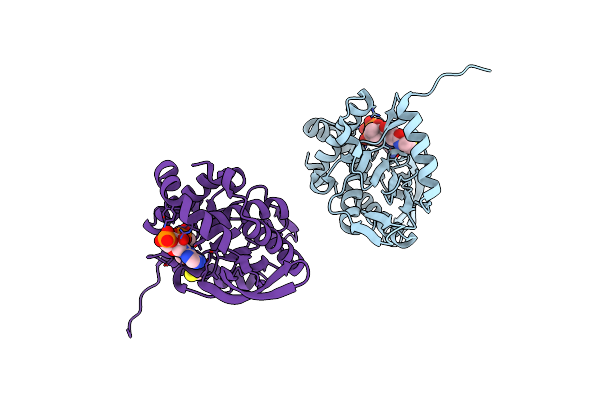 |
Crystal Structure Of The Catalytic Domain Of Corynebacterium Mustelae Predicted Acetyltransferase Acef (E2P).
Organism: Corynebacterium mustelae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-08-18 Classification: TRANSFERASE Ligands: COA |
 |
Organism: Anoxybacillus flavithermus (strain dsm 21510 / wk1)
Method: ELECTRON MICROSCOPY Release Date: 2020-08-12 Classification: MEMBRANE PROTEIN Ligands: PTY, K |
 |
Organism: Streptomyces lincolnensis
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2020-01-15 Classification: TRANSFERASE |
 |
Organism: Streptomyces lincolnensis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-01-15 Classification: TRANSFERASE |
 |
Organism: Staphylococcus phage k
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-12-20 Classification: VIRAL PROTEIN Ligands: ACT, NA |
 |
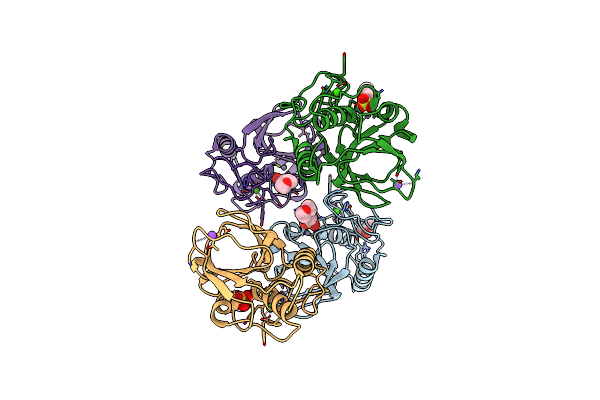
Alcohol Dehydrogenase From The Antarctic Psychrophile Moraxella Sp. Tae 123
Organism: Moraxella sp. (strain tae123)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-04-13 Classification: OXIDOREDUCTASE Ligands: ZN |
 |
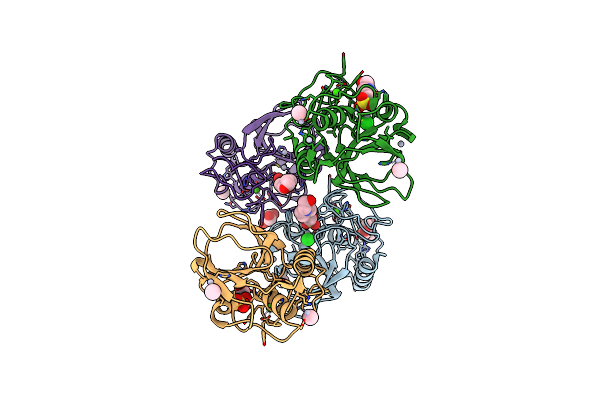
Native Structure Of The Lytic Chapk Domain Of The Endolysin Lysk From Staphylococcus Aureus Bacteriophage K
Organism: Staphylococcus phage k
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2014-08-06 Classification: HYDROLASE Ligands: GOL, CA, MES, NA, ZN |
 |
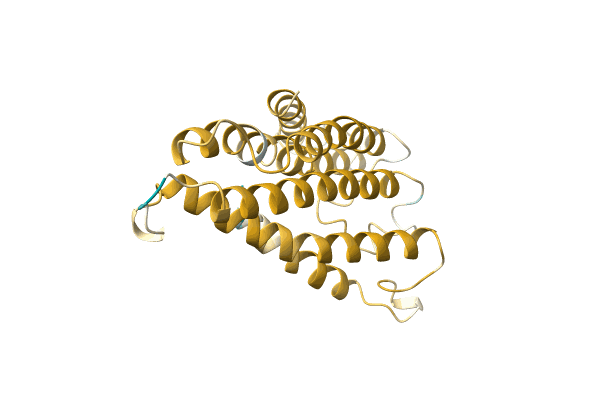
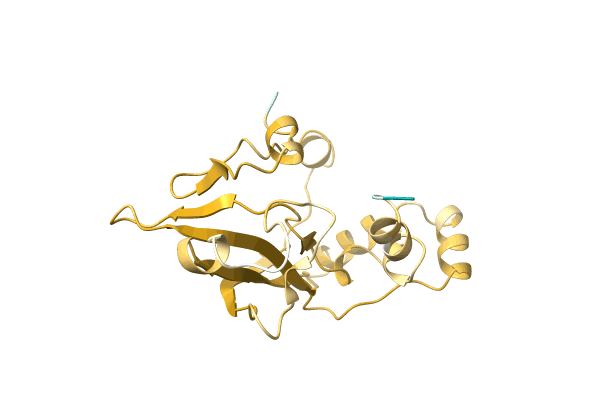
Methylmercury Chloride Derivative Structure Of The Lytic Chapk Domain Of The Endolysin Lysk From Staphylococcus Aureus Bacteriophage K
Organism: Kayvirus kay
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2014-08-06 Classification: VIRAL PROTEIN Ligands: CL, GOL, EPE, CA, MMC, HG |
 |
 |
 |
 |
 |
 |

