Search Count: 81
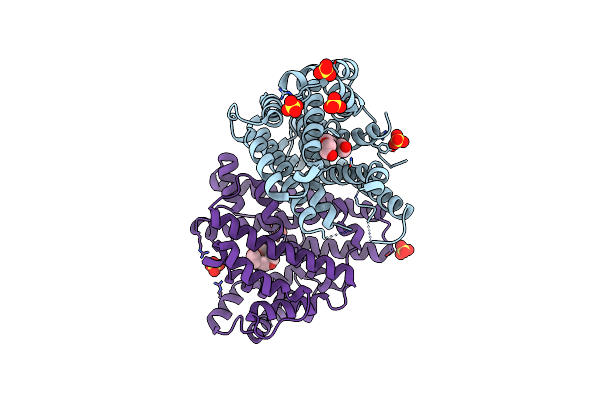 |
Crystal Structure Of Pentalenene Synthase Variant F76A With Peg Molecule In The Active Site
Organism: Streptomyces exfoliatus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-11-06 Classification: METAL BINDING PROTEIN Ligands: PG4, SO4 |
 |
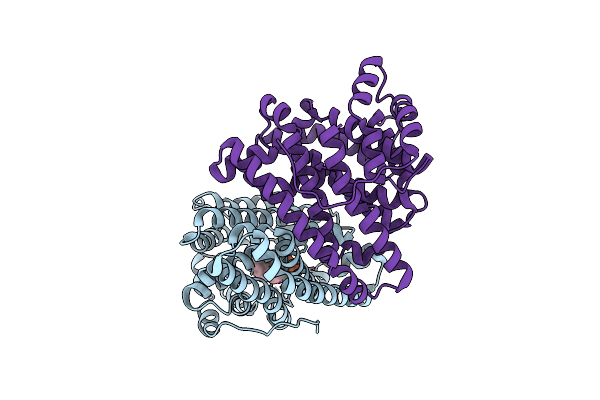
Crystal Structure Of Pentalenene Synthase Variant F76A Complexed With 2-Fluorofarnesyl Diphosphate
Organism: Streptomyces exfoliatus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-11-06 Classification: METAL BINDING PROTEIN Ligands: FPF, MG |
 |
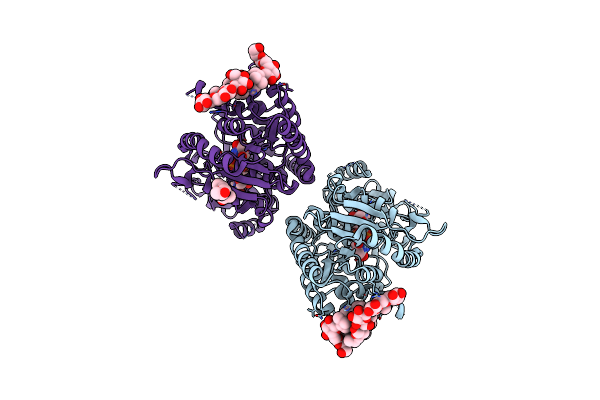
Crystal Structure Of Pentalenene Synthase Variant F76A Complexed With 12,13-Difluorofarnesyl Diphosphate
Organism: Streptomyces exfoliatus
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-11-06 Classification: METAL BINDING PROTEIN Ligands: FDF, MG |
 |
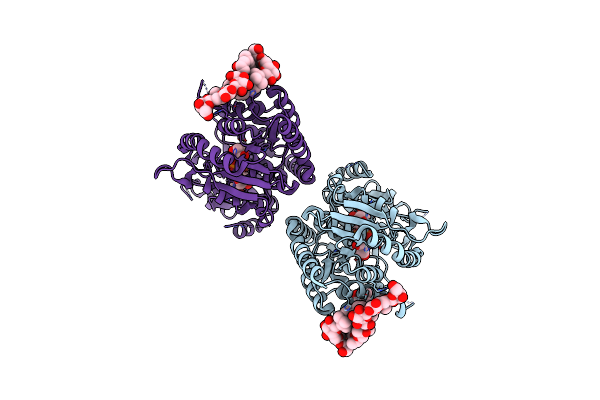
Organism: Siraitia grosvenorii
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2024-09-04 Classification: TRANSFERASE Ligands: UD1 |
 |
Crystal Structure Of Glycosyltransferase Sgugt94-289-3 In Complex With M5, State 2
Organism: Siraitia grosvenorii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-05-29 Classification: TRANSFERASE Ligands: M1K, POG, TRS, UDP |
 |
Crystal Structure Of Glycosyltransferase Sgugt94-289-3 In Complex With M5, State 1
Organism: Siraitia grosvenorii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-05-29 Classification: TRANSFERASE Ligands: POG, M1K, UDP, TRS |
 |
Crystal Structure Of Glycosyltransferase Sgugt94-289-3 In Complex With Sia, State 3
Organism: Siraitia grosvenorii
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2024-05-29 Classification: TRANSFERASE Ligands: POG, M0O, UDP, TRS |
 |
Crystal Structure Of Glycosyltransferase Sgugt94-289-3 In Complex With Sia, State 1
Organism: Siraitia grosvenorii
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2024-05-29 Classification: TRANSFERASE Ligands: M0O, UDP |
 |
Organism: Siraitia grosvenorii
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2024-05-29 Classification: TRANSFERASE Ligands: UDP, TRS, POG, M7F |
 |
Organism: Siraitia grosvenorii
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2024-05-29 Classification: TRANSFERASE Ligands: UPG |
 |
Crystal Structure Of Glycosyltransferase Sgugt94-289-3 In Complex With Udp State 2
Organism: Siraitia grosvenorii
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2024-05-29 Classification: TRANSFERASE Ligands: UDP, TRS, PEG |
 |
Crystal Structure Of Glycosyltransferase Sgugt94-289-3 In Complex With Udp State 1
Organism: Siraitia grosvenorii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-05-29 Classification: TRANSFERASE Ligands: POG, UDP |
 |
Organism: Siraitia grosvenorii
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2024-05-29 Classification: TRANSFERASE |
 |
Crystal Structure Of Glycosyltransferase Sgugt94-289-3 In Complex With M3, State 2
Organism: Siraitia grosvenorii
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2024-05-29 Classification: TRANSFERASE Ligands: UDP, M2Q |
 |
Crystal Structure Of A Ugt Transferase From Siraitia Grosvenorii In Complex With Udp
Organism: Siraitia grosvenorii
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-03-03 Classification: TRANSFERASE Ligands: UDP |
 |
Organism: Siraitia grosvenorii
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-04-08 Classification: TRANSFERASE |
 |
Organism: Siraitia grosvenorii
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2020-04-01 Classification: TRANSFERASE |
 |
Organism: Siraitia grosvenorii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-04-01 Classification: TRANSFERASE Ligands: UPG |
 |
Organism: Siraitia grosvenorii
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2020-04-01 Classification: TRANSFERASE Ligands: SO4 |
 |
Crystal Structure Of Cqsb2 From Streptomyces Exfoliatus 2419-Svt2 (Apo Form)
Organism: Streptomyces exfoliatus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-10-16 Classification: BIOSYNTHETIC PROTEIN |

