Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN Ligands: F42 |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN Ligands: F42 |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN Ligands: F42 |
 |
Organism: Streptomyces
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Streptomyces
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Streptomyces
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2024-11-13 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Crystal Structure Of O-Carbamoyltransferase Vtdb And The Compound Vtdb With Carbamoyladenylate From Streptomyces Sp. No1W98
Organism: Streptomyces
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2024-01-24 Classification: TRANSFERASE Ligands: MG |
 |
Crystal Structure Of Cytochrome P450 Aspb Bound To N1-Methylated Cyclo-L-Trp-L-Pro
Organism: Streptomyces
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2023-09-20 Classification: OXIDOREDUCTASE Ligands: HEM, RQB, ZN |
 |
Crystal Structure Of The O-Carbamoyltransferase Vtdb In Complex With Carbamoyl Adenylate Intermediate
Organism: Streptomyces
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2023-03-22 Classification: TRANSFERASE Ligands: MG, CA0 |
 |
Organism: Streptomyces
Method: ELECTRON MICROSCOPY Release Date: 2022-11-09 Classification: PROTEIN BINDING |
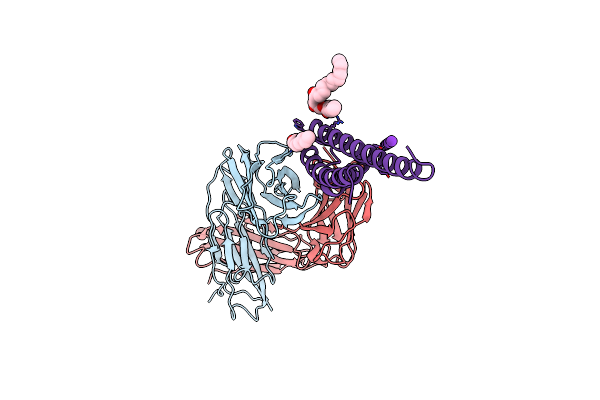 |
Structure Of The Kcsa-W67F Mutant With The Activation Gate In The Closed Conformation
Organism: Mus musculus, Streptomyces
Method: X-RAY DIFFRACTION Resolution:3.21 Å Release Date: 2022-10-12 Classification: MEMBRANE PROTEIN Ligands: F09, 1EM, K |
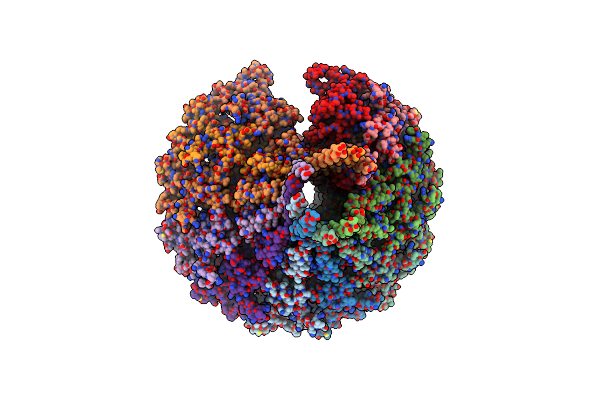 |
Organism: Pseudomonas virus pap3
Method: ELECTRON MICROSCOPY Release Date: 2022-04-20 Classification: VIRAL PROTEIN |
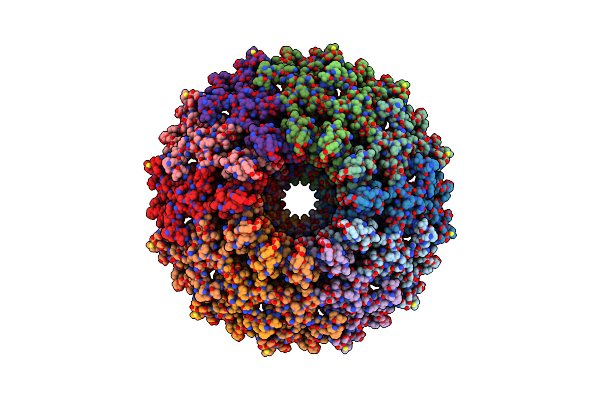 |
Organism: Pseudomonas virus pap3
Method: ELECTRON MICROSCOPY Release Date: 2022-04-20 Classification: VIRAL PROTEIN |
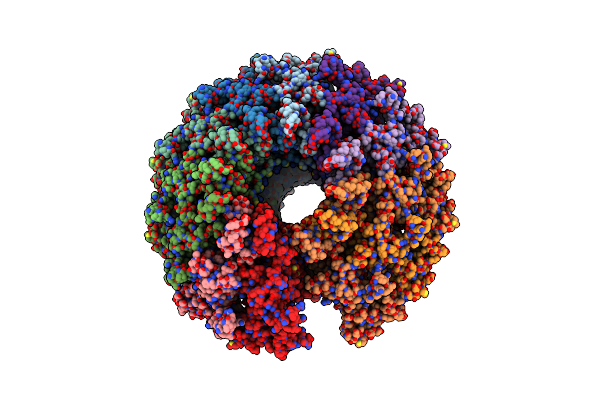 |
Kinetically Trapped Pseudomonas-Phage Pap3 Portal Protein - Delta Barrel Mutant Class-2
Organism: Pseudomonas virus pap3
Method: ELECTRON MICROSCOPY Release Date: 2022-03-30 Classification: VIRAL PROTEIN |
 |
Kinetically Trapped Pseudomonas-Phage Pap3 Portal Protein - Delta Barrel Mutant Class-3
Organism: Pseudomonas virus pap3
Method: ELECTRON MICROSCOPY Release Date: 2022-03-30 Classification: VIRAL PROTEIN |
 |
Organism: Actinobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2022-03-02 Classification: HYDROLASE Ligands: TRS, MG |
 |
Organism: Actinobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-10-13 Classification: HYDROLASE Ligands: IOD |