Search Count: 3,533
 |
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: DNA BINDING PROTEIN |
 |
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: FLAVOPROTEIN Ligands: FMN |
 |
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: FLAVOPROTEIN Ligands: FAD |
 |
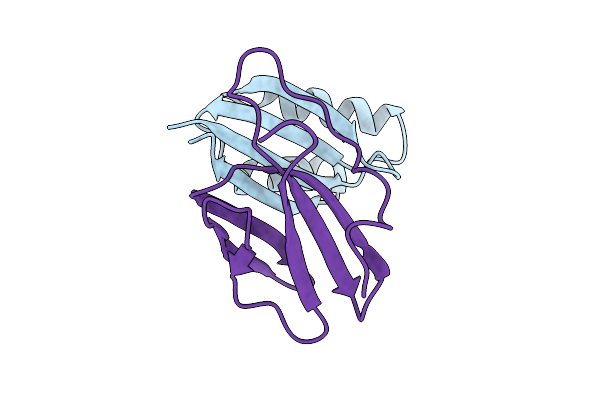
Complex Of Rice Blast (Magnaporthe Oryzae) Effector Protein Avr-Pia With The Hma Domain Of Oshpp09 From Rice (Oryza Sativa)
Organism: Oryza sativa, Pyricularia oryzae
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: PLANT PROTEIN Ligands: SO4 |
 |
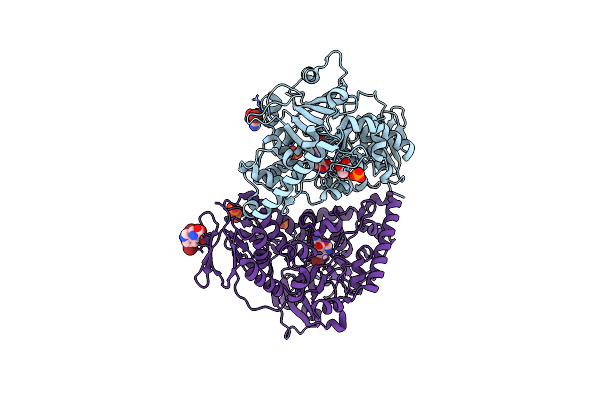
The Complex Of Rice Immune Receptor Rga5-Hma8 With Rice Blast Effector Protein Avr-Pikd
Organism: Oryza sativa, Pyricularia oryzae
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: PLANT PROTEIN |
 |
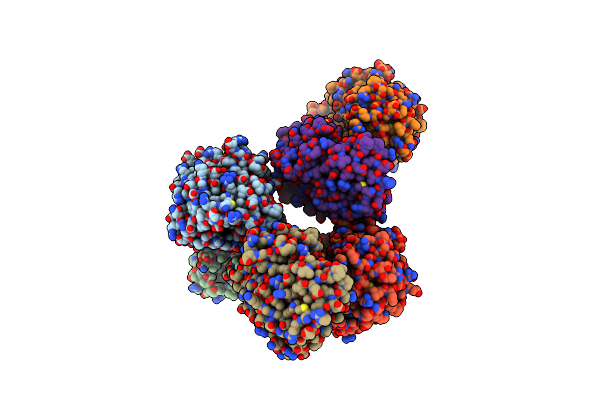
Flavin-Dependent Tryptophan 6-Halogenase Thal In Complex With 6-Bromo-L-Tryptophan
Organism: Streptomyces albogriseolus
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: FLAVOPROTEIN Ligands: BTR, PO4, GOL |
 |
Organism: Kitasatospora sp.
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4, UNL |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN Ligands: F42 |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN Ligands: F42 |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN Ligands: F42 |
 |
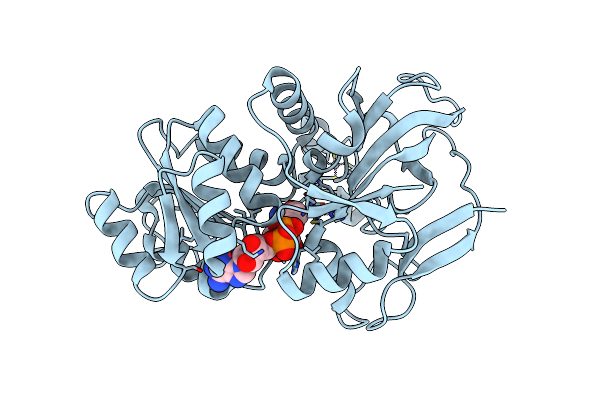
Organism: Streptomyces albogriseolus
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: FLAVOPROTEIN Ligands: WYH, GOL, PO4, NA |
 |
Organism: Modestobacter roseus
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2025-04-23 Classification: OXIDOREDUCTASE Ligands: NAD, ZN |
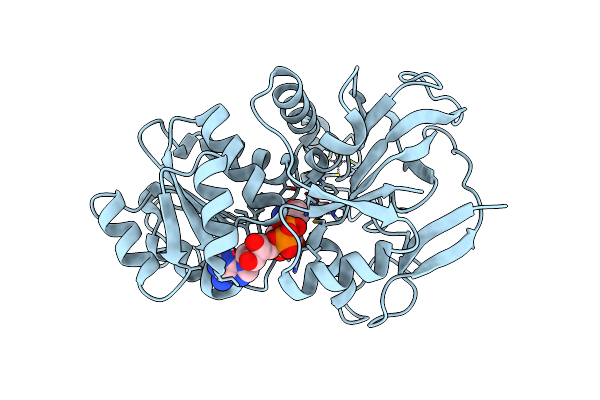 |
Organism: Modestobacter roseus
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2025-04-23 Classification: OXIDOREDUCTASE Ligands: NAD, ZN |
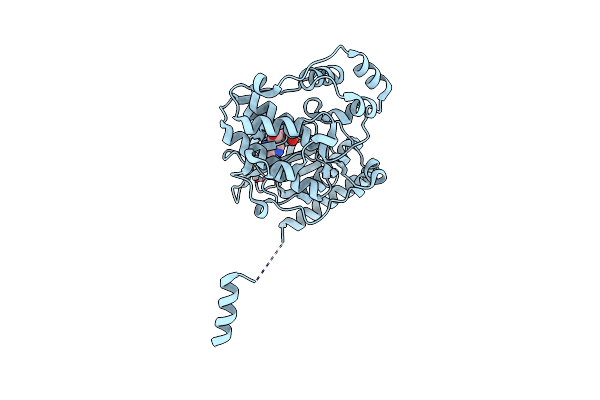 |
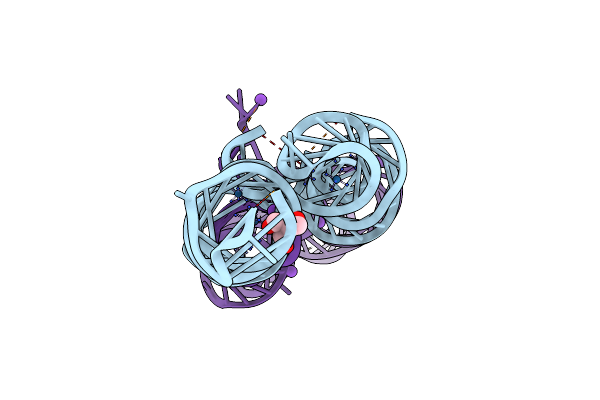
Organism: Pyricularia oryzae
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2025-04-23 Classification: SIGNALING PROTEIN Ligands: AMP |
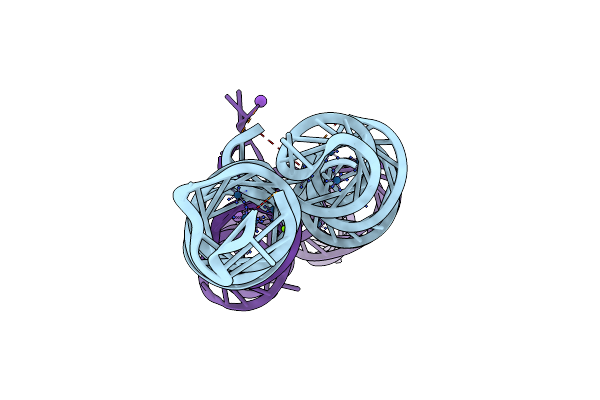 |
Organism: Rous sarcoma virus - prague c
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2025-04-09 Classification: RNA Ligands: IRI, MG, K |
 |
Organism: Rous sarcoma virus - prague c
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-04-09 Classification: RNA Ligands: IRI, K, TAM |
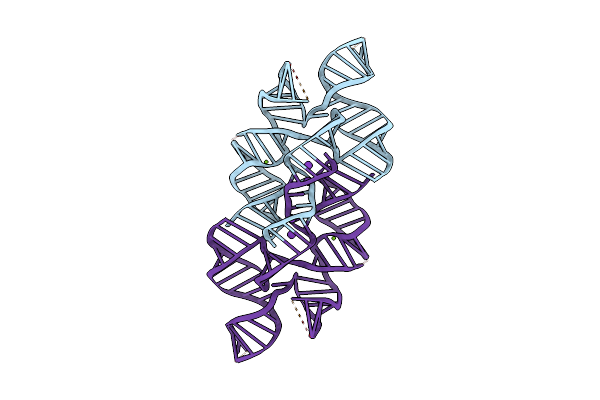 |
Organism: Rous sarcoma virus - prague c
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: RNA Ligands: K, MG |
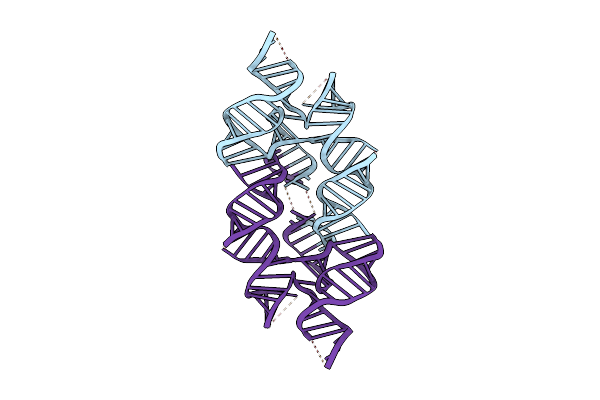 |
Organism: Rous sarcoma virus - prague c
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: RNA |

