Search Count: 611
 |
Organism: Rhodococcus sp. (in: high g+c gram-positive bacteria)
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: LIGASE Ligands: CL, A1JC1 |
 |
Organism: Rhodococcus sp. (in: high g+c gram-positive bacteria)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-06-05 Classification: HYDROLASE Ligands: XSR, CL |
 |
Cryo-Em Structure Of A Bacterial Nitrilase Filament With A Covalent Adduct Derived From Benzonitrile Hydrolysis
Organism: Rhodococcus sp. (in: high g+c gram-positive bacteria)
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: HYDROLASE Ligands: HBX |
 |
Organism: Rhodococcus sp. (in: high g+c gram-positive bacteria)
Method: X-RAY DIFFRACTION Release Date: 2024-04-17 Classification: HYDROLASE Ligands: TN9, CL, SO4 |
 |
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7 Circular Permutated At Positions 141-156 (Cphalotagdelta)
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-10-11 Classification: HYDROLASE Ligands: NHE |
 |
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7 Circular Permutated At Positions 141-156 (Cphalotagdelta) Fused To M13
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-10-11 Classification: HYDROLASE Ligands: CL |
 |
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7 Circular Permutated At Positions 154-156 (Cphalotag7_154-156)
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2023-10-11 Classification: HYDROLASE Ligands: CL |
 |
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7 With An Insertion Of Calmodulin-M13 Fusion At Position 154-156 That Mimic The Structure Of Caprola, An Calcium Gated Protein Labeling Technology
Organism: Rhodococcus sp. (in: high g+c gram-positive bacteria), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-10-11 Classification: HYDROLASE Ligands: CL, CA |
 |
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7 Labeled With A Chloroalkane Cyanine3 Fluorophore Substrate
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-07-26 Classification: HYDROLASE Ligands: PJI, CL, GOL, MG |
 |
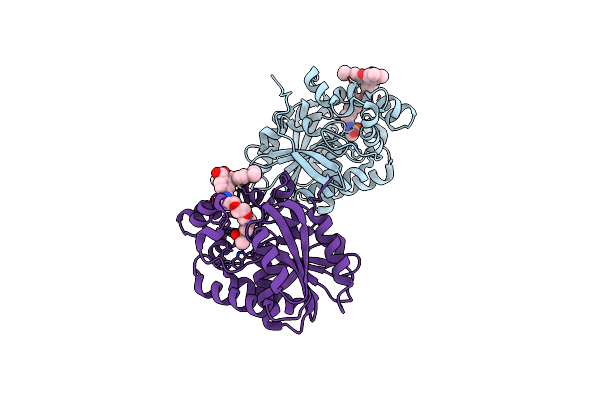
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7 Fusion To The Green Fluorescent Protein Gfp (Chemog1) Labeled With A Chloroalkane Tetramethylrhodamine Fluorophore Substrate
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-07-26 Classification: HYDROLASE Ligands: OEH, CL, GOL |
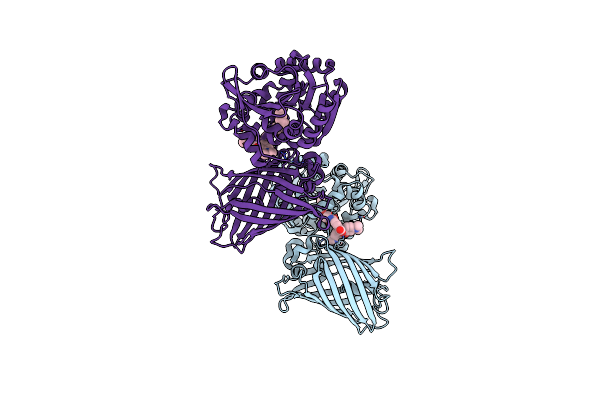 |
X-Ray Structure Of The Interface Optimized Haloalkane Dehalogenase Halotag7 Fusion To The Green Fluorescent Protein Gfp (Chemog5-Tmr) Labeled With A Chloroalkane Tetramethylrhodamine Fluorophore Substrate
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-07-26 Classification: HYDROLASE Ligands: OEH, CL |
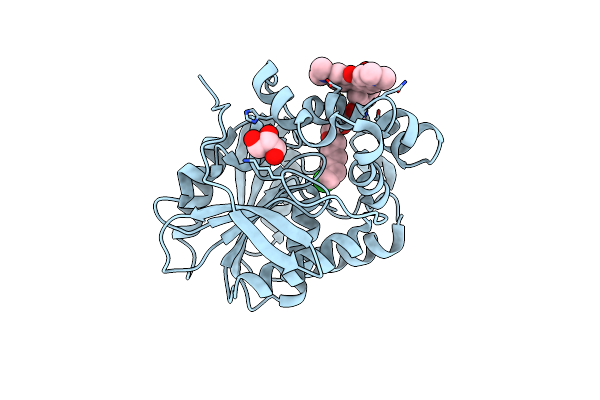 |
X-Ray Structure Of The Haloalkane Dehalogenase Dead Variant Halotag7-D106A Bound To A Chloroalkane Tetramethylrhodamine Fluorophore Ligand (Ca-Tmr)
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2023-04-19 Classification: HYDROLASE Ligands: OEH, CL, GOL |
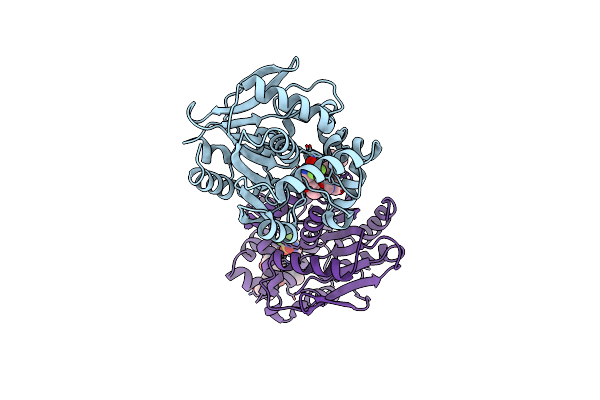 |
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7 Bound To A Butyltrifluoromethanesulfonamide Tetramethylrhodamine Ligand (Tmr-T4)
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2023-04-19 Classification: HYDROLASE Ligands: IYE |
 |
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7 Bound To A Butylmethanesulfonamide Tetramethylrhodamine Ligand (Tmr-S4)
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2023-04-19 Classification: HYDROLASE Ligands: IYU |
 |
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7 Bound To A Pentyltrifluoromethanesulfonamide Tetramethylrhodamine Ligand (Tmr-T5)
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-02-22 Classification: HYDROLASE Ligands: IYI, CA |
 |
X-Ray Structure Of The Dead Variant Haloalkane Dehalogenase Halotag7-D106A Bound To A Pentanol Tetramethylrhodamine Ligand (Tmr-Hy5)
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-02-22 Classification: HYDROLASE Ligands: IYL, CL, GOL |
 |
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7 Bound To A Pentylmethanesulfonamide Tetramethylrhodamine Ligand (Tmr-S5)
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-02-22 Classification: HYDROLASE Ligands: IYO, GOL |
 |
Cryo-Em Structure Of The T=4 Lake Sinai Virus 2 Virus-Like Capsid At Ph 7.5
Organism: Lake sinai virus 2
Method: ELECTRON MICROSCOPY Release Date: 2023-02-08 Classification: VIRUS LIKE PARTICLE |
 |
Cryo-Em Structure Of The T=3 Lake Sinai Virus 2 Virus-Like Capsid At Ph 7.5
Organism: Lake sinai virus 2
Method: ELECTRON MICROSCOPY Release Date: 2023-02-08 Classification: VIRUS LIKE PARTICLE |
 |
Cryo-Em Structure Of The T=4 Lake Sinai Virus 2 Virus-Like Capsid At Ph 6.5
Organism: Lake sinai virus 2
Method: ELECTRON MICROSCOPY Release Date: 2023-02-08 Classification: VIRUS LIKE PARTICLE |

