Search Count: 443
 |
Organism: Streptomyces sp. ms191
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: FE2 |
 |
Organism: Rhodococcus
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2025-02-26 Classification: OXIDOREDUCTASE |
 |
Organism: Oceanobacillus iheyensis
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: RNA Ligands: MG |
 |
Organism: Oceanobacillus iheyensis
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: RNA Ligands: MG |
 |
Organism: Septifer virgatus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-12-02 Classification: SUGAR BINDING PROTEIN Ligands: CL |
 |
Organism: Septifer virgatus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-12-02 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Streptococcus phage 73
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-06-06 Classification: VIRAL PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2014-05-14 Classification: UNKNOWN FUNCTION |
 |
Organism: Oceanobacillus iheyensis
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2010-02-02 Classification: HYDROLASE Ligands: CA, CL, EPE |
 |
Crystal Structure Of Nucleoside Diphosphate Kinase From Haloarcula Quadrata
Organism: Haloarcula quadrata
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2009-08-25 Classification: TRANSFERASE |
 |
Organism: Rhodococcus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2009-08-18 Classification: ELECTRON TRANSPORT Ligands: HEM |
 |
Organism: Rhodococcus
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2009-08-18 Classification: ELECTRON TRANSPORT Ligands: IMD, EDO, HEM |
 |
Crystal Structure Of Divergent Enolase From Oceanobacillus Iheyensis Complexed With Mg And L-Malate.
Organism: Oceanobacillus iheyensis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-10-21 Classification: Isomerase, lyase Ligands: LMR, MG |
 |
Crystal Structure Of Divergent Enolase From Oceanobacillus Iheyensis Complexed With Mg And L-Malate.
Organism: Oceanobacillus iheyensis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-10-21 Classification: Isomerase, lyase Ligands: LMR, MG |
 |
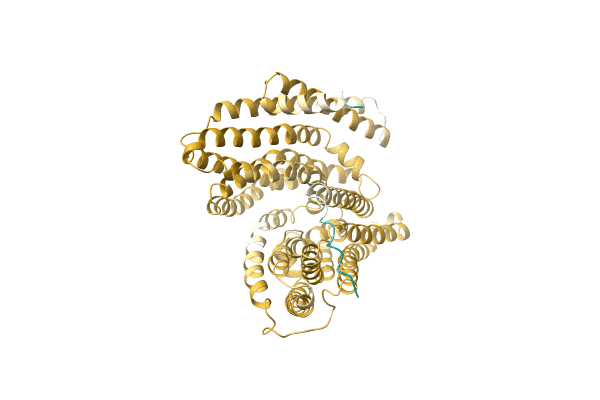
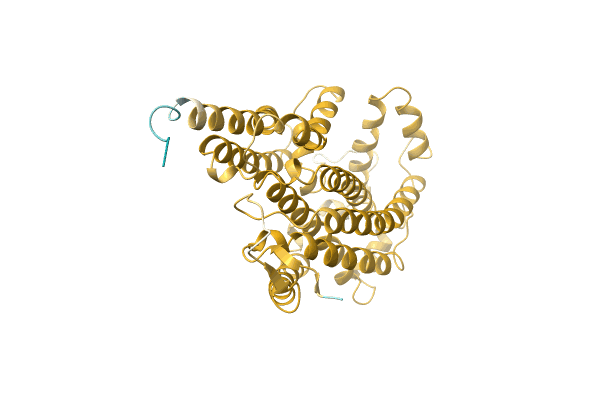
The Crystal Structure Of Muconate Cycloisomerase From Oceanobacillus Iheyensis
Organism: Oceanobacillus iheyensis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-03-06 Classification: ISOMERASE Ligands: MG |
 |
 |
 |
 |
 |

