Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
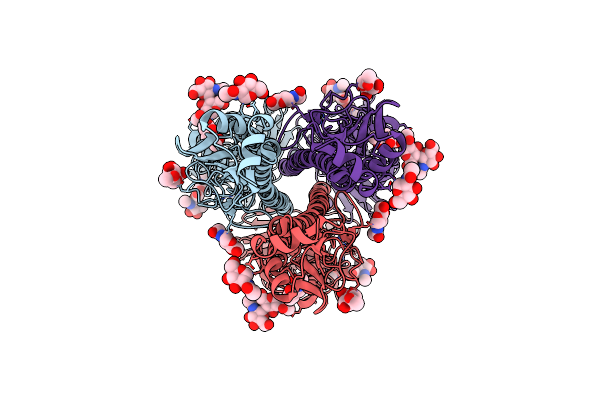
Crystal Structure Of Gdx-Clo From Small Multidrug Resistance Family Of Transporters In Low Ph (Protonated State)
Organism: Clostridiales bacterium oral taxon 876, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2022-03-02 Classification: TRANSPORT PROTEIN Ligands: 9PD |
 |
Organism: Influenza a virus (a/michigan/45/2015(h1n1))
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2021-12-08 Classification: HYDROLASE Ligands: NAG, CA |
 |
Organism: Pseudomonas phage jbd16c
Method: SOLUTION NMR Release Date: 2021-05-26 Classification: VIRAL PROTEIN |
 |
Organism: Pseudomonas phage jbd16c
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2021-05-19 Classification: IMMUNE SYSTEM |
 |
Localized Reconstruction Of The H1 A/Michigan/45/2015 Ectodomain Displayed At The Surface Of I53_Dn5 Nanoparticle
Organism: Influenza a virus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-03-31 Classification: VIRAL PROTEIN Ligands: NAG |
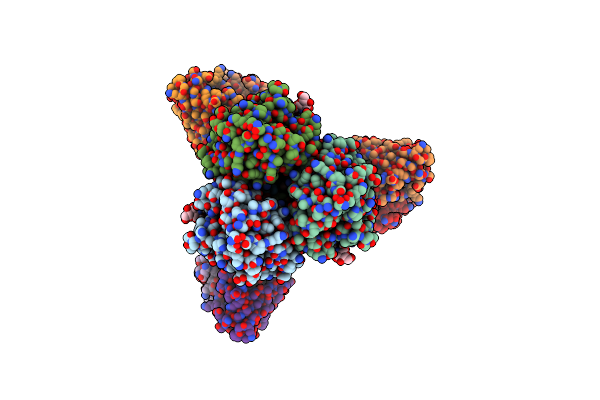 |
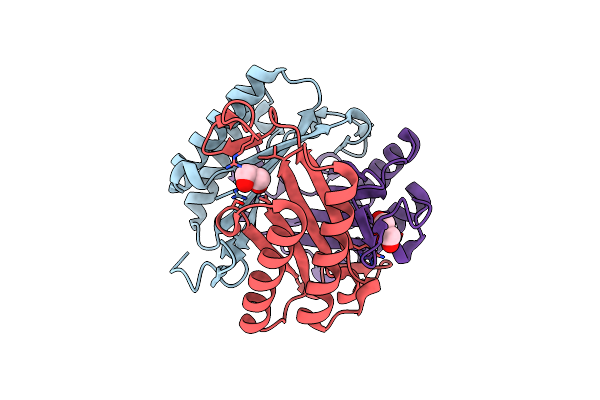
Cryoem Structure Of Influenza Hemagglutinin A/Michigan/45/2015 In Complex With Cyno Antibody 1C4
Organism: Influenza a virus (a/michigan/45/2015(h1n1)), Macaca fascicularis
Method: ELECTRON MICROSCOPY Release Date: 2021-03-10 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
 |
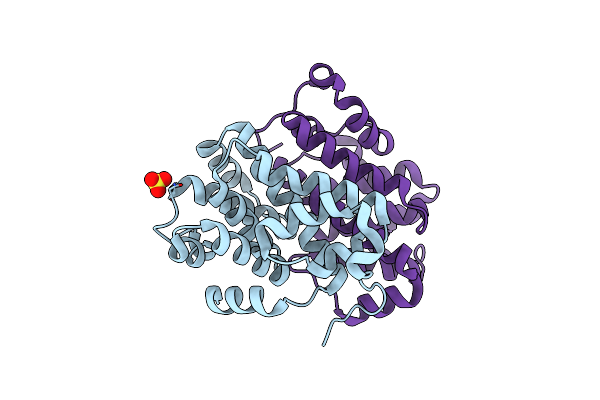
Organism: Caballeronia arationis
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2020-04-15 Classification: ISOMERASE Ligands: GOL |
 |
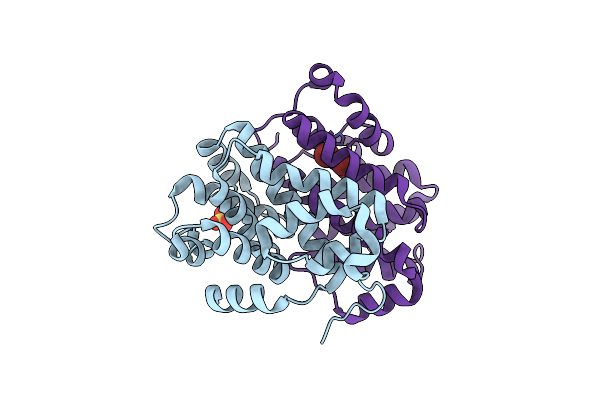
Organism: Marinomonas mediterranea (strain atcc 700492 / jcm 21426 / nbrc 103028 / mmb-1)
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2019-05-29 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4 |
 |
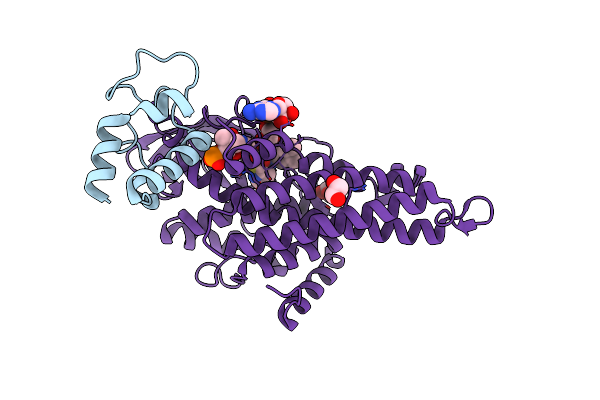
Crystal Structure Of The Debrominase Bmp8 C82A In Complex With 2,3,4-Tribromopyrrole
Organism: Marinomonas mediterranea mmb-1
Method: X-RAY DIFFRACTION Resolution:3.19 Å Release Date: 2019-05-29 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4, MKV |
 |
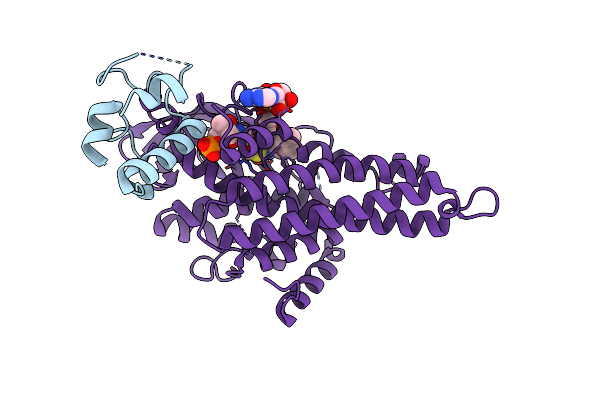
Organism: Marinomonas mediterranea (strain atcc 700492 / jcm 21426 / nbrc 103028 / mmb-1)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-01-16 Classification: OXIDOREDUCTASE Ligands: FK4, FAD, EDO |
 |
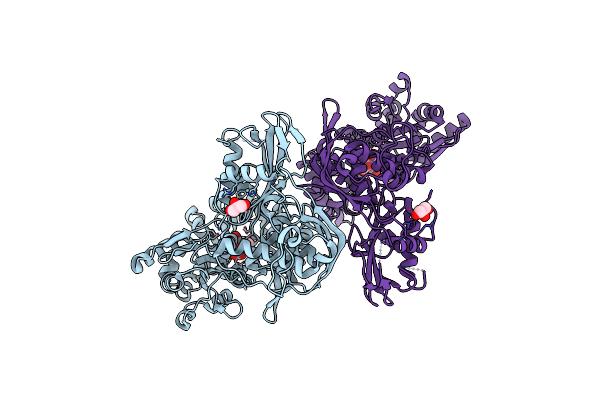
Organism: Marinomonas mediterranea (strain atcc 700492 / jcm 21426 / nbrc 103028 / mmb-1)
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2019-01-16 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD, PNS |
 |
Crystal Structure Of The Cas6 Domain Of Marinomonas Mediterranea Mmb-1 Cas6-Rt-Cas1 Fusion Protein
Organism: Escherichia coli o157:h7, Marinomonas mediterranea mmb-1
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2018-10-17 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
Organism: Marinomonas mediterranea (strain atcc 700492 / jcm 21426 / nbrc 103028 / mmb-1)
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2018-02-07 Classification: ISOMERASE Ligands: PO4 |
 |
Organism: Marinomonas mediterranea
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2013-10-23 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of The L-Lys Epsilon-Oxidase From Marinomonas Mediterranea
Organism: Marinomonas mediterranea
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2013-09-04 Classification: OXIDOREDUCTASE Ligands: SO4, DIO, EDO |
 |
Crystal Structure Of The Schiff Base Intermediate Of L-Lys Epsilon-Oxidase From Marinomonas Mediterranea With L-Lys
Organism: Marinomonas mediterranea
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2013-09-04 Classification: OXIDOREDUCTASE Ligands: LYS, SO4, EDO, DIO |
 |
Organism: Mycobacterium gastri
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-11-03 Classification: LYASE Ligands: CL, MG, SO4 |
 |
 |
 |