Search Count: 571
 |
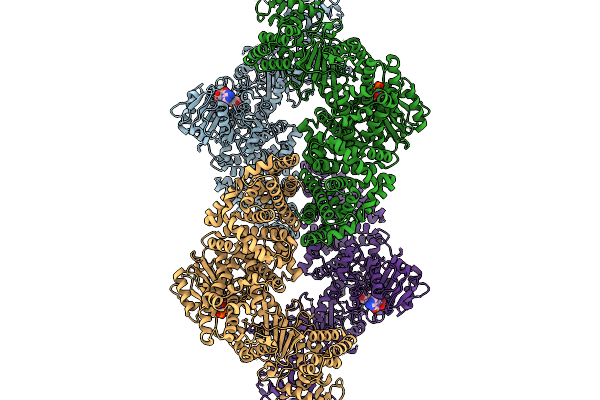 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Open Tetramer.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
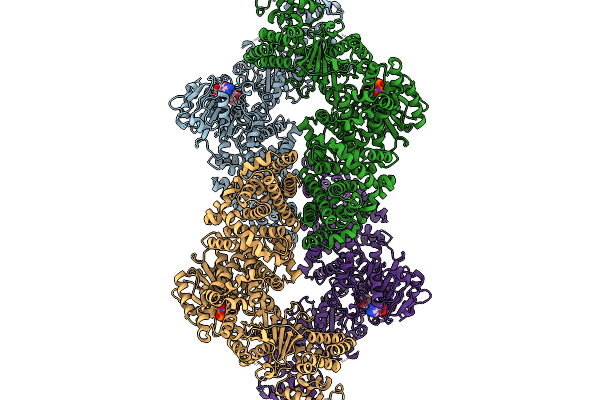 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Closed1 Tetramer.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
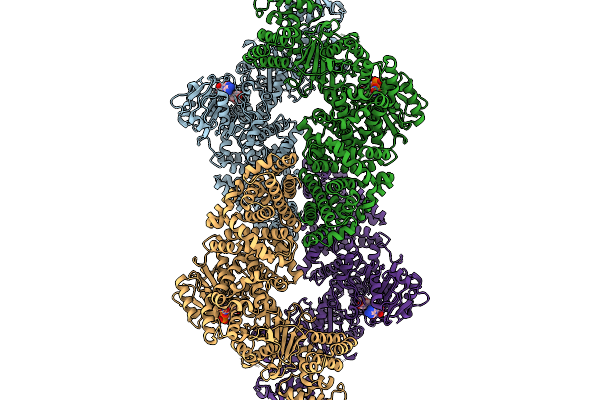 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Closed2 Tetramer
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Empty Monomer.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE |
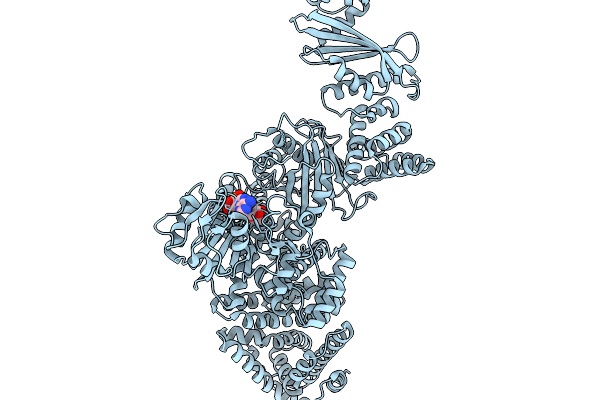 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Cofactor-Monomer.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Cofactor/Ligand-Monomer
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: GLU, NAD |
 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Total-Monomer
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Organism: Mycolicibacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: A1I1V, RFP, DMS |
 |
Organism: Mycolicibacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-08-27 Classification: OXIDOREDUCTASE Ligands: IMP |
 |
Crystal Structure Of M. Smegmatis Gmp Reductase With Xmp* Intermidiate In Complex With Nadp+ And Imp.
Organism: Mycolicibacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-08-27 Classification: OXIDOREDUCTASE Ligands: IMP, NAP |
 |
Crystal Structure Of M. Smegmatis Gmp Reductase In Complex With Gmp And Atp.
Organism: Mycolicibacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-08-27 Classification: OXIDOREDUCTASE Ligands: ATP, 5GP |
 |
Crystal Structure Of M. Smegmatis Gmp Reductase In Complex With Imp And Atp.
Organism: Mycolicibacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-08-27 Classification: OXIDOREDUCTASE Ligands: IMP, ATP |
 |
Crystal Structure Of M. Smegmatis Gmp Reductase In Complex With Gmp And Gtp.
Organism: Mycolicibacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-08-27 Classification: OXIDOREDUCTASE Ligands: 5GP, GTP |
 |
Cryoem Structure Of M. Smegmatis Gmp Reductase In Complex With Gmp At Ph 6.6, Extended Conformation.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: 5GP |
 |
Cryoem Structure Of M. Smegmatis Gmp Reductase In Complex With Gmp At Ph 6.6, Compressed Conformation.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: 5GP |
 |
Cryoem Structure Of M. Smegmatis Gmp Reductase In Complex With Gmp And Gtp At Ph 6.6, Extended Conformation I.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: 5GP, GTP |
 |
Cryoem Structure Of M. Smegmatis Gmp Reductase In Complex With Gmp And Gtp At Ph 6.6, Extended Conformation Ii.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: 5GP, GTP |
 |
Cryoem Structure Of M. Smegmatis Gmp Reductase In Complex With Gmp And Atp At Ph 6.6, Compressed Conformation.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: 5GP, ATP |
 |
Cryoem Structure Of M. Smegmatis Gmp Reductase Apoform At Ph 6.6, Extended Conformation I.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: OXIDOREDUCTASE |

