Search Count: 32
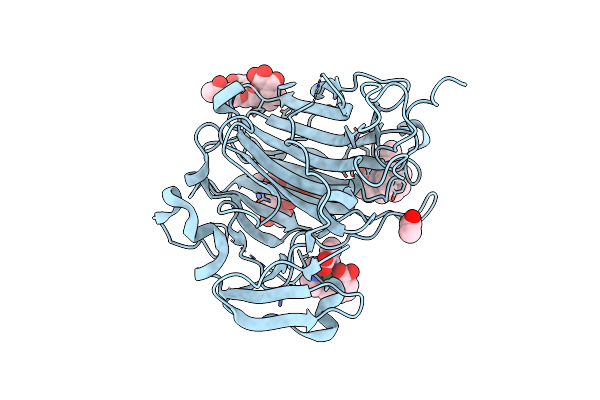 |
Crystal Structure Of The Zymogen Form Of The Glutamic-Class Prolyl-Endopeptidase Neprosin At 2.05 A Resolution In Presence Of The Crystallophore Lu-Xo4.
Organism: Nepenthes ventricosa x nepenthes alata
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-08-10 Classification: HYDROLASE Ligands: K93, ACT |
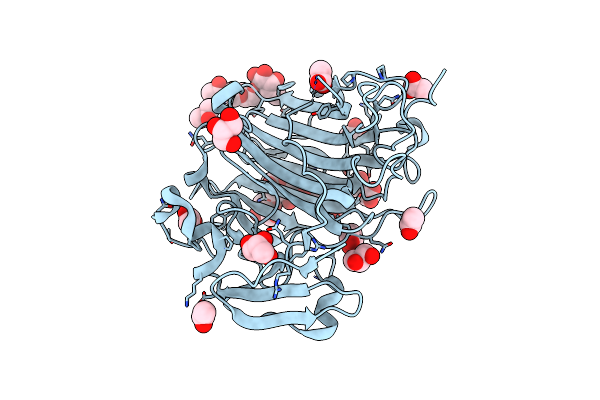 |
Crystal Structure Of The Native Zymogen Form Of The Glutamic-Class Prolyl-Endopeptidase Neprosin At 1.80 A Resolution.
Organism: Nepenthes ventricosa x nepenthes alata
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-08-10 Classification: HYDROLASE Ligands: GOL, ACT, IPA |
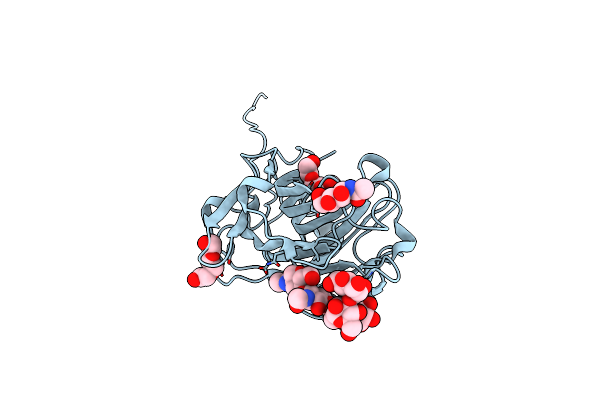 |
Crystal Structure Of The Mature Form Of The Glutamic-Class Prolyl-Endopeptidase Neprosin At 2.35 A Resolution.
Organism: Nepenthes ventricosa x nepenthes alata
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2022-08-10 Classification: HYDROLASE Ligands: NAG, PGE |
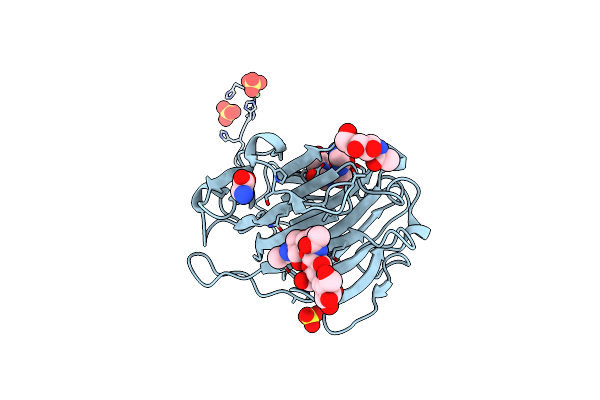 |
Second Crystal Form Of The Mature Glutamic-Class Prolyl-Endopeptidase Neprosin At 1.85 A Resolution.
Organism: Nepenthes ventricosa x nepenthes alata
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-08-10 Classification: HYDROLASE Ligands: NAG, SO4, NI, GLY |
 |
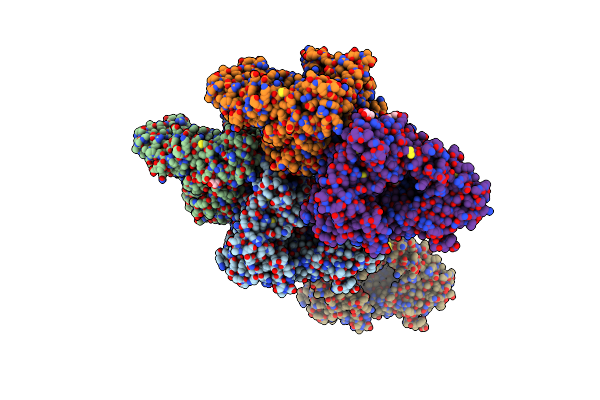
Crystal Structure Of L182E D-Succinylase (Dsa) From Cupriavidus Sp. P4-10-C
Organism: Cupriavidus sp. p4-10-c
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-01-26 Classification: HYDROLASE Ligands: CAD, GOL, SIN, CO3, CA, CL |
 |
Organism: Cupriavidus sp. p4-10-c
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-01-26 Classification: HYDROLASE Ligands: GOL, CO3, CA |
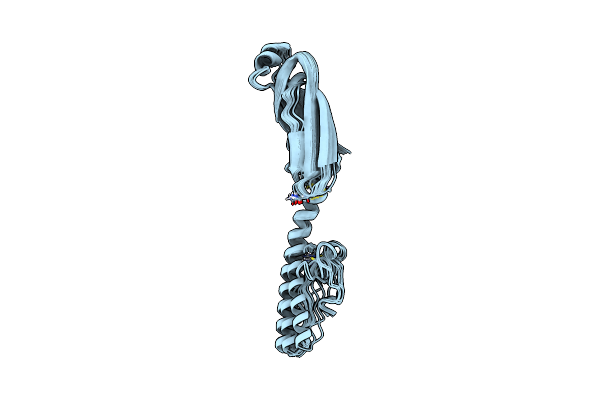 |
Full Structure Of Rymv P1 Protein, Derived From Crystallographic And Nmr Data.
Organism: Rice yellow mottle virus
Method: SOLUTION NMR Release Date: 2021-02-03 Classification: VIRAL PROTEIN Ligands: ZN |
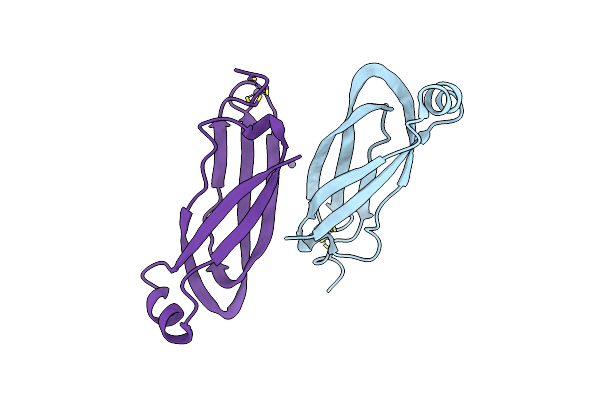 |
Nt Part Crystal Structure Of The Rymv-Encoded Viral Rna Silencing Suppressor P1
Organism: Rice yellow mottle virus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-01-27 Classification: RNA BINDING PROTEIN Ligands: ZN |
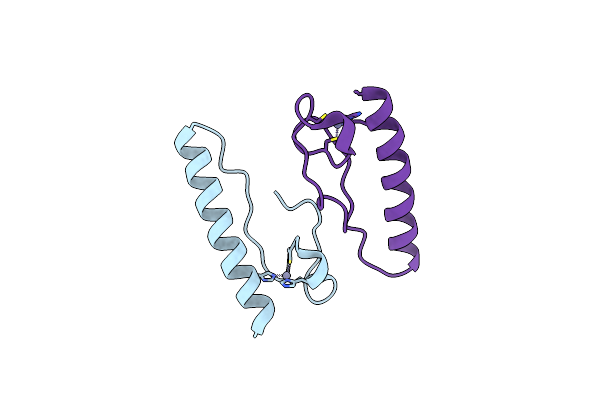 |
Ct Part Crystal Structure Of The Rymv-Encoded Viral Rna Silencing Suppressor P1
Organism: Rice yellow mottle virus
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2021-01-27 Classification: RNA BINDING PROTEIN Ligands: ZN |
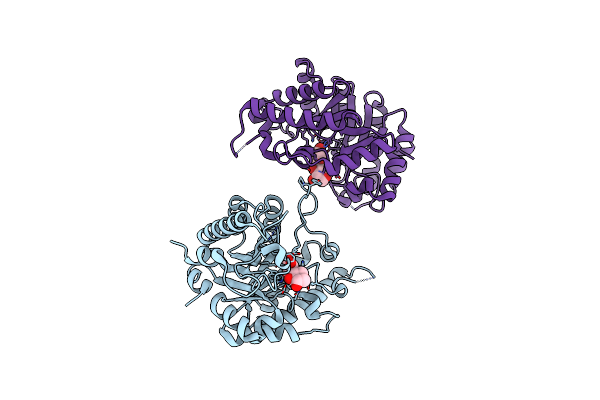 |
Organism: Environmental samples
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-02-24 Classification: HYDROLASE |
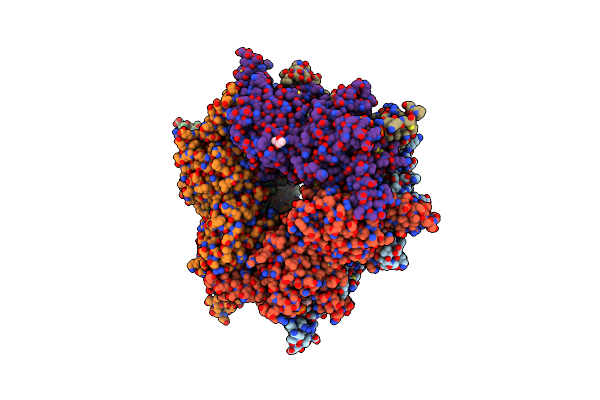 |
Transcarboxylase 12S Crystal Structure: Hexamer Assembly And Substrate Binding To A Multienzyme Core (With Methylmalonyl-Coenzyme A And Methylmalonic Acid Bound)
Organism: Propionibacterium freudenreichii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2003-05-20 Classification: TRANSFERASE Ligands: CD, MCA, DXX, MPD |
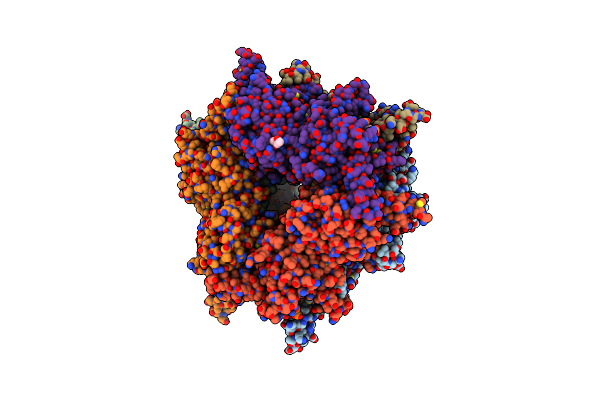 |
Transcarboxylase 12S Crystal Structure: Hexamer Assembly And Substrate Binding To A Multienzyme Core (With Hydrolyzed Methylmalonyl-Coenzyme A Bound)
Organism: Propionibacterium freudenreichii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-05-20 Classification: TRANSFERASE Ligands: CD, MCA, MPD |
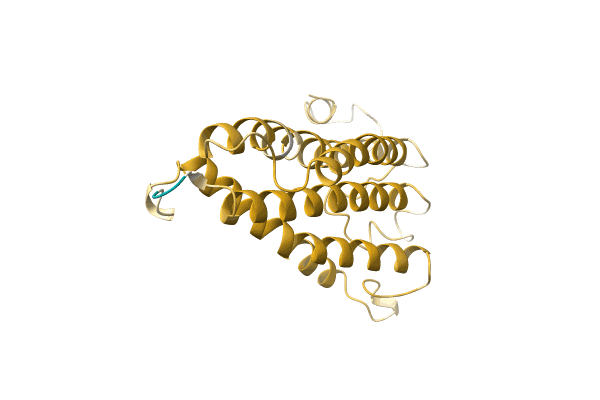 |
 |
Organism: Scrobipalpopsis sp. BOLD:AAG9147
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Propionibacterium freudenreichii
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Propionibacterium freudenreichii
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Propionibacterium freudenreichii
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Propionibacterium freudenreichii
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Propionibacterium freudenreichii
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Propionibacterium freudenreichii
Method: Alphafold Release Date: Classification: NA Ligands: NA |

