Search Count: 41,488
 |
Organism: Corynebacterium glutamicum
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSPORT PROTEIN Ligands: ABU, PGT, CDL |
 |
Organism: Corynebacterium glutamicum
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSPORT PROTEIN Ligands: PGT, ABU, LMT |
 |
Organism: Corynebacterium glutamicum
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSPORT PROTEIN Ligands: PGT, BET, NA |
 |
Organism: Mycolicibacterium smegmatis
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: A1I1V, RFP, DMS |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: A1BK4 |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: A1BK3, GOL |
 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Substrate-Bound State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
 |
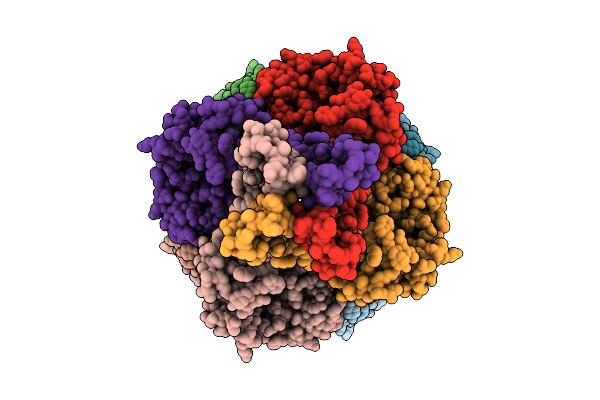
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Pre-Catalysis And Product-Bound State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG, UDP, A1B94 |
 |
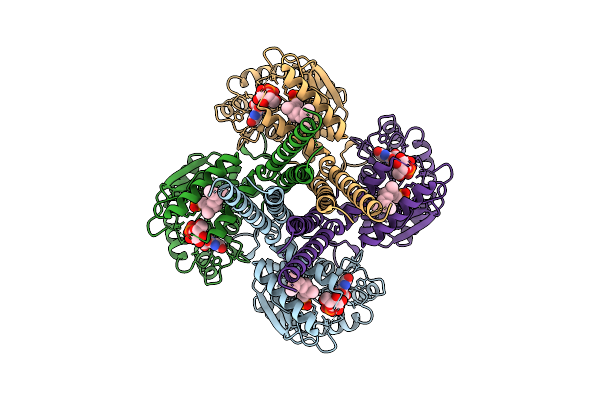
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Apo State (Octamer Volume)
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
 |
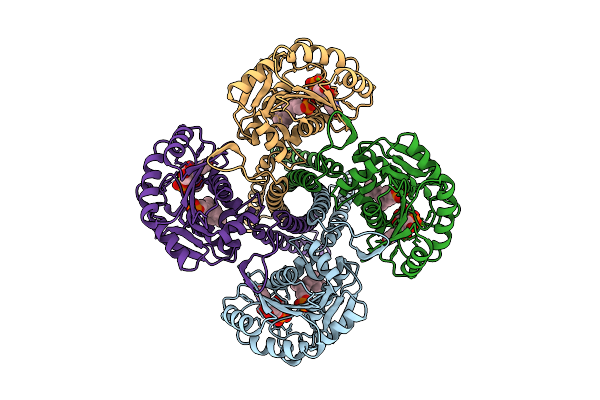
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
 |
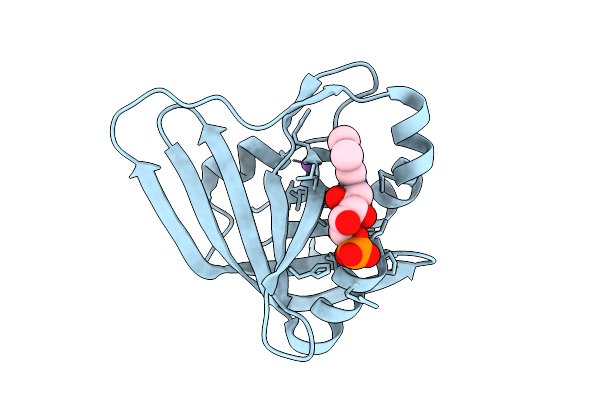
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Pre-Intermediate State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
 |
Organism: Mycolicibacterium fortuitum
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: FLAVOPROTEIN |
 |
Organism: Mycolicibacterium fortuitum
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: FLAVOPROTEIN |
 |
M. Tuberculosis Clpc1-Ntd Complexed With A Click Chemistry Analog Of Rufomycin
Organism: Mycobacterium tuberculosis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: ANTIBIOTIC |
 |
Organism: Mycobacterium tuberculosis, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN |
 |
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: FMN |
 |
Structure Of Acp Domain Conjugated With Stearic Acid And Interacting With Mpt Domain From M. Tuberculosis Type-I Fas
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Simian immunodeficiency virus, Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN Ligands: ZN |
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM |
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, ARG |

