Search Count: 110
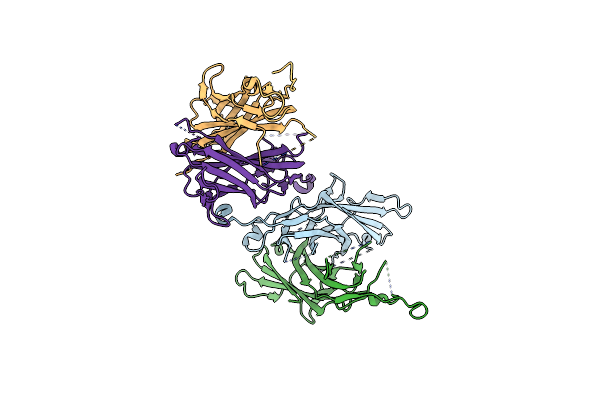 |
Organism: Aspergillus nidulans
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-05-07 Classification: CELL CYCLE |
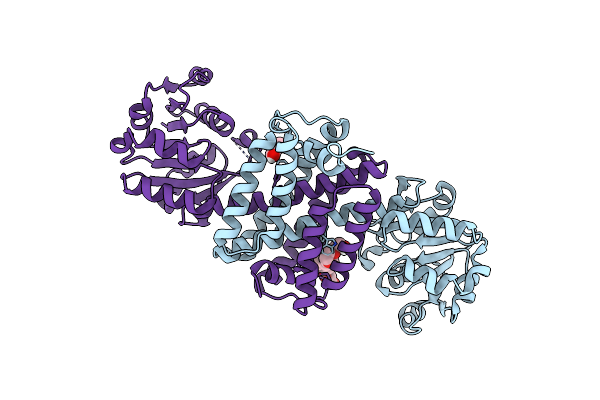 |
Organism: Paenibacillus mucilaginosus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-08-21 Classification: OXIDOREDUCTASE Ligands: PEG, P4C |
 |
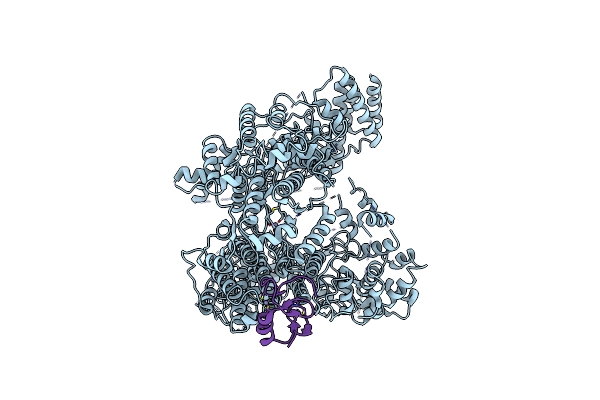
Lymphocytic Choriomeningitis Virus Rna-Dependent Rna Polymerase (Lcmv-L Protein)
Organism: Lymphocytic choriomeningitis virus (strain armstrong)
Method: ELECTRON MICROSCOPY Release Date: 2023-09-13 Classification: TRANSFERASE Ligands: MN |
 |
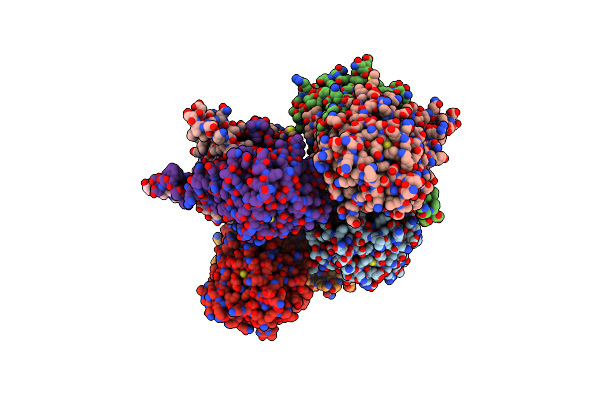
Lymphocytic Choriomeningitis Virus Polymerase- Matrix Z Protein Complex (Lcmv L-Z Complex)
Organism: Lymphocytic choriomeningitis virus (strain armstrong)
Method: ELECTRON MICROSCOPY Release Date: 2023-09-13 Classification: METAL BINDING PROTEIN/TRANSFERASE Ligands: ZN, MN |
 |
Structure Of Lactobacillus Salivarius (Ls) Bile Salt Hydrolase(Bsh) In Complex With Glycocholate (Gca)
Organism: Ligilactobacillus salivarius
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-03-08 Classification: HYDROLASE Ligands: GCH |
 |
Structure Of Lactobacillus Salivarius (Ls) Bile Salt Hydrolase(Bsh) In Complex With Taurocholate (Tca)
Organism: Ligilactobacillus salivarius
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-03-08 Classification: HYDROLASE Ligands: TCH |
 |
Organism: Paenibacillus mucilaginosus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-02-22 Classification: OXIDOREDUCTASE |
 |
Organism: Nephila antipodiana
Method: SOLUTION NMR Release Date: 2021-11-17 Classification: PROTEIN FIBRIL |
 |
N-Terminal Domain (Ntd) Solution Structure Of Aciniform Spidroin (Acspn) From Nephila Antipodiana.
Organism: Nephila antipodiana
Method: SOLUTION NMR Release Date: 2020-07-22 Classification: STRUCTURAL PROTEIN |
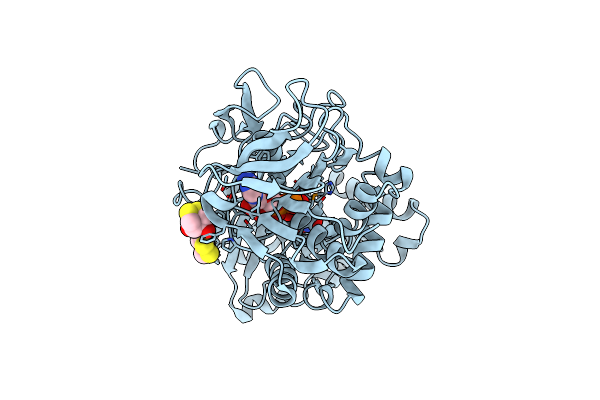 |
Crystal Structure Of The Modified Fructosyl Peptide Oxidase From Aspergillus Nidulans, Seleno-Methionine Derivative
Organism: Aspergillus nidulans
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2019-05-15 Classification: OXIDOREDUCTASE Ligands: FAD, D1D, SO4 |
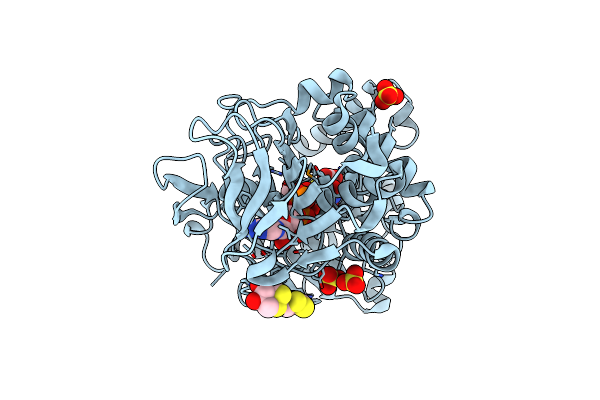 |
Crystal Structure Of The Modified Fructosyl Peptide Oxidase From Aspergillus Nidulans In Complex With Fsa, Seleno-Methionine Derivative
Organism: Aspergillus nidulans
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-05-15 Classification: OXIDOREDUCTASE Ligands: FAD, FSA, D1D, SO4 |
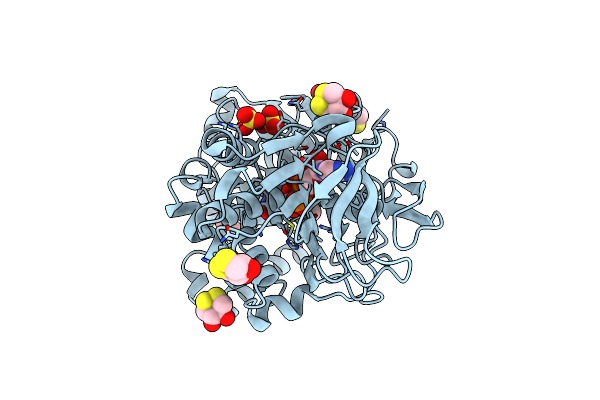 |
Crystal Structure Of The Modified Fructosyl Peptide Oxidase From Aspergillus Nidulans With R61G Mutation
Organism: Aspergillus nidulans
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-05-15 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, D1D |
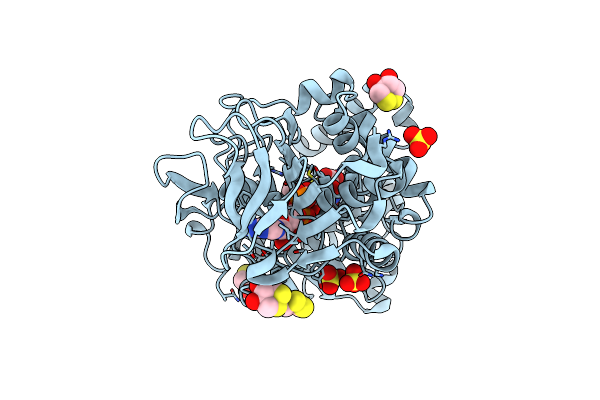 |
Crystal Structure Of The Modified Fructosyl Peptide Oxidase From Aspergillus Nidulans With R61G Mutation, In Complex With Fsa
Organism: Aspergillus nidulans
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-05-15 Classification: OXIDOREDUCTASE Ligands: FAD, FSA, D1D, SO4 |
 |
Crystal Structure Of The Modified Fructosyl Peptide Oxidase From Aspergillus Nidulans With 7 Additional Mutations, In Complex With Fsa
Organism: Aspergillus nidulans
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2019-05-15 Classification: OXIDOREDUCTASE Ligands: FAD, FSA |
 |
Bile Salt Hydrolase From Lactobacillus Salivarius Complex With Glycocholic Acid And Cholic Acid
Organism: Lactobacillus salivarius
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-08-22 Classification: HYDROLASE Ligands: GCH, CHD, GOL, PO4, EDO, PEG |
 |
Organism: Lactobacillus salivarius
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-05-11 Classification: HYDROLASE Ligands: PO4 |
 |
Organism: Nephila antipodiana
Method: SOLUTION NMR Release Date: 2015-06-17 Classification: STRUCTURAL PROTEIN |
 |
2.8A Crystal Structure Of Lymphocytic Choriomeningitis Virus Nucleoprotein C-Terminal Domain
Organism: Lymphocytic choriomeningitis virus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-06-11 Classification: HYDROLASE Ligands: ZN, IMD, MG, K |
 |
2.0A Crystal Structure Of Lymphocytic Choriomeningitis Virus Nucleoprotein C-Terminal Domain
Organism: Lymphocytic choriomeningitis virus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-06-11 Classification: HYDROLASE Ligands: ZN, MG, IMD |
 |
Untangling The Solution Structure Of C-Terminal Domain Of Aciniform Spidroin
Organism: Nephila antipodiana
Method: SOLUTION NMR Release Date: 2014-03-05 Classification: STRUCTURAL PROTEIN |

