Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Crystal Structure Of Glucose 1-Dehydrogenase Mutant1 From Limosilactobacillus Fermentum
Organism: Limosilactobacillus fermentum
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2024-10-30 Classification: HYDROLASE |
 |
Crystal Structure Of Glucose 1-Dehydrogenase Mutant2 From Limosilactobacillus Fermentum
Organism: Limosilactobacillus fermentum
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2024-10-30 Classification: HYDROLASE |
 |
Crystal Structure Of Glucose 1-Dehydrogenase From Limosilactobacillus Fermentum
Organism: Limosilactobacillus fermentum
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-10-30 Classification: HYDROLASE Ligands: MG |
 |
Crystal Structure Of A Short-Chain Dehydrogenase From Lactobacillus Fermentum With Nadph
Organism: Limosilactobacillus fermentum
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-10-30 Classification: HYDROLASE Ligands: NAP |
 |
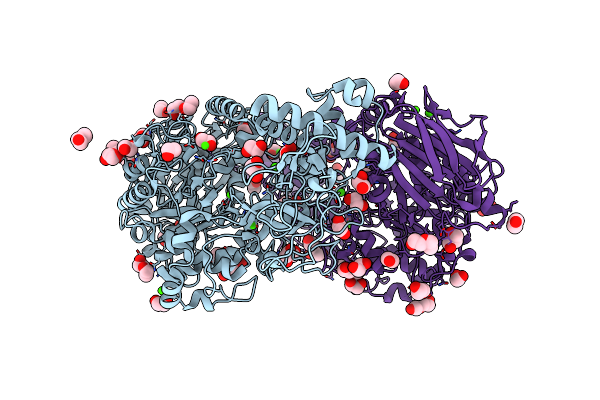
Conversion Of Indole-3-Acetic Acid Into Indole-3-Aldehyde In Bacteria Metabolic Network Of Tryptophan Around The Indole-3-Aldehyde Formation
Organism: Limosilactobacillus fermentum
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2023-07-05 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
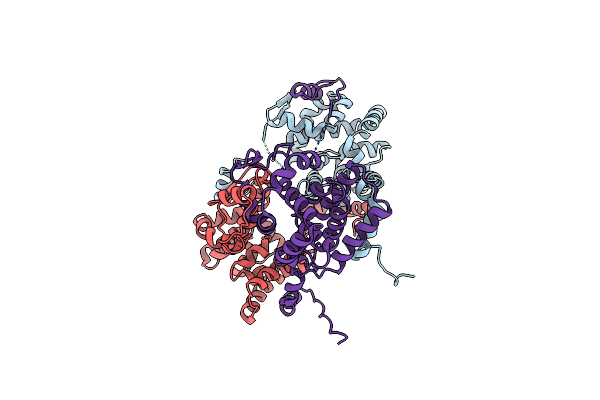
Organism: Limosilactobacillus fermentum
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2022-01-12 Classification: TRANSFERASE Ligands: CA, EDO, GOL, PEG |
 |
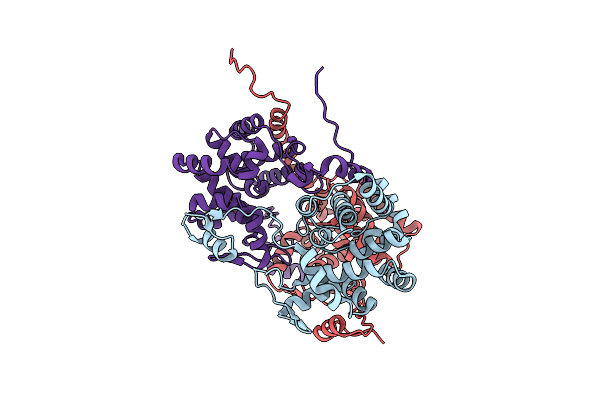
Structure Of Tomato Spotted Wilt Virus Nucleocapsid Protein With Alternative Oligomerization State
Organism: Tomato spotted wilt virus
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2017-09-27 Classification: VIRAL PROTEIN |
 |
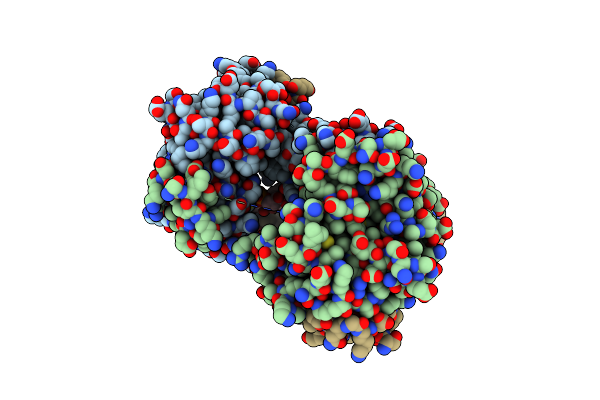
Organism: Tomato spotted wilt virus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-03-22 Classification: VIRAL PROTEIN |
 |
Organism: Tomato spotted wilt virus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2017-03-22 Classification: VIRAL PROTEIN/RNA |
 |
Organism: Tomato spotted wilt virus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2017-03-22 Classification: Viral protein/DNA |
 |
Organism: Escherichia coli (strain k12), Escherichia coli mp020980.2, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.99 Å Release Date: 2016-06-22 Classification: TRANSCRIPTION/DNA |
 |
Crystal Structure Of A Putative Mandelate Racemase/Muconate Lactonizing Enzyme Family Protein From Roseovarius
Organism: Roseovarius sp. tm1035
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2011-08-17 Classification: ISOMERASE Ligands: SO4, FMT, GOL |
 |
Crystal Structure Of Mandelate Racemase/Muconate Lactonizing Enzyme-Like Protein From Roseovarius Sp. Tm1035
Organism: Roseovarius sp. tm1035
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2011-05-18 Classification: ISOMERASE Ligands: MG, GOL, RIB |
 |
 |
Organism: Hypaurotis quercus (purple hairstreak)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Hypaurotis quercus (purple hairstreak)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Hypaurotis quercus (purple hairstreak)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
 |
 |