Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Comparative Analysis Of Functions And Catalytic Mechanisms Of Methyltransferases Involved In Anthracycline Biosynthesis
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: TRANSFERASE Ligands: SAH, A1L3V |
 |
Comparative Analysis Of Functions And Catalytic Mechanisms Of Methyltransferases Involved In Anthracycline Biosynthesis
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: TRANSFERASE Ligands: A1EI6, SAH |
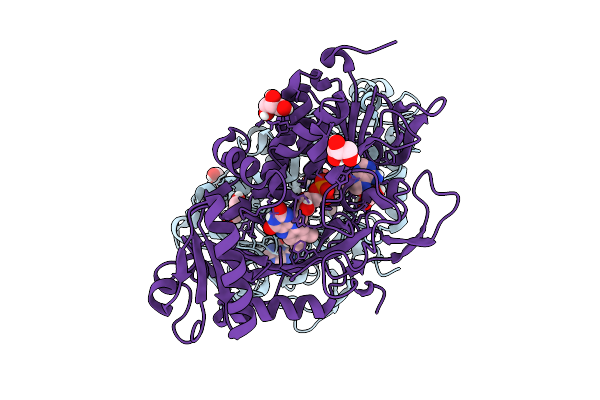 |
Organism: Parastagonospora nodorum sn15
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: FAD, GOL |
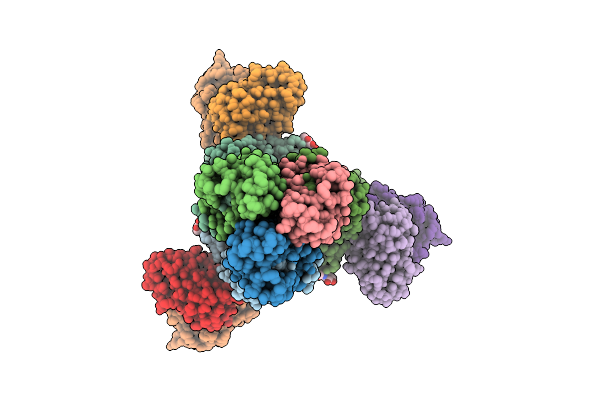 |
Cryoem Structure Of H7 Hemagglutinin In Complex With A Human Neutralizing Antibody 6Y13
Organism: Influenza a virus (a/duck/chiba/25-51-14/2013(h7n1)), Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: VIRAL PROTEIN Ligands: NAG |
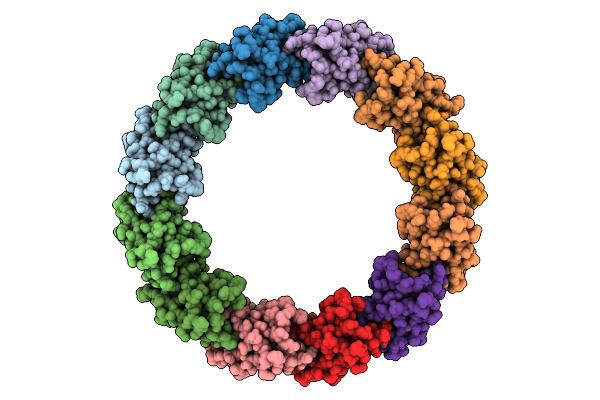 |
Organism: Escherichia phage lambda
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: VIRAL PROTEIN |
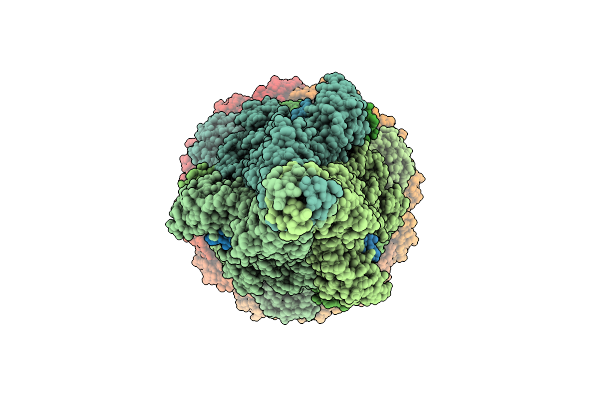 |
Organism: Escherichia phage lambda
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: VIRAL PROTEIN Ligands: SF4 |
 |
The Bifunctional Arabinofuranosidase/Xylosidase From Metagenome Of Pseudacanthotermes Militaris.
Organism: Pseudacanthotermes militaris
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: CA, TRS, GOL |
 |
Distinct Quaternary States, Intermediates, And Autoinhibition During Loading Of The Dnab-Replicative Helicase By The Phage Lambda P Helicase Loader
Organism: Escherichia phage lambda
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: REPLICATION |
 |
Ecoli Dnab Helicase And Phage Lambda Loader P With Adp-Mg In A 6:5 Stoichiometry Ratio.
Organism: Escherichia coli, Escherichia phage lambda
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: DNA BINDING PROTEIN Ligands: ADP, MG |
 |
Ecoli Dnab Helicase And Phage Lambda Loader P With Adp-Mg In A 6:6 Stoichiometry Ratio.
Organism: Escherichia coli, Escherichia phage lambda
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: DNA BINDING PROTEIN Ligands: ADP, MG |
 |
Crystal Structure Of An Inverse Charged Cutinase Mutant From Saccharopolyspora Flava (611)
Organism: Saccharopolyspora flava
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2025-04-16 Classification: HYDROLASE |
 |
Organism: Peltigera membranacea, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-04-02 Classification: DNA BINDING PROTEIN Ligands: ANP, MG |
 |
Organism: Peltigera membranacea, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-04-02 Classification: DNA BINDING PROTEIN Ligands: ANP, MG |
 |
Organism: Pseudomonas rhizosphaerae
Method: X-RAY DIFFRACTION Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: HEM, OXY, CL, SO4 |
 |
Crystal Structure Of Fncas12A In Complex With Pre-Crrna And 12Nt Target Dna
Organism: Francisella tularensis subsp. novicida u112
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-01 Classification: RNA BINDING PROTEIN Ligands: MG |
 |
Crystal Structure Of Fncas12A In Complex With Pre-Crrna And 18Nt Target Dna
Organism: Francisella tularensis subsp. novicida u112
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2025-01-01 Classification: RNA BINDING PROTEIN Ligands: MG |
 |
Crystal Structure Of Adenosylcobinamide Kinase / Adenosylcobinamide Phosphate Guanylyltransferase From Methylocapsa Palsarum
Organism: Methylocapsa palsarum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-01 Classification: TRANSFERASE Ligands: PO4 |
 |
Crystal Structure Of Adenosylcobinamide Kinase / Adenosylcobinamide Phosphate Guanylyltransferase Complexed With Adenosylcobinamide-Phosphate
Organism: Methylocapsa palsarum
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-01-01 Classification: TRANSFERASE Ligands: B12, 5AD, PO4 |
 |
Adenosylcobinamide Kinase/Adenosylcobinamide Phosphate Guanylyltransferase -C91S
Organism: Methylocapsa palsarum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-01-01 Classification: TRANSFERASE Ligands: PO4 |
 |
Crystal Structure Of Adenosylcobinamide Kinase / Adenosylcobinamide Phosphate Guanylyltransferase Complexed With Gdp
Organism: Methylocapsa palsarum
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2025-01-01 Classification: TRANSFERASE Ligands: GDP, DPO |