Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
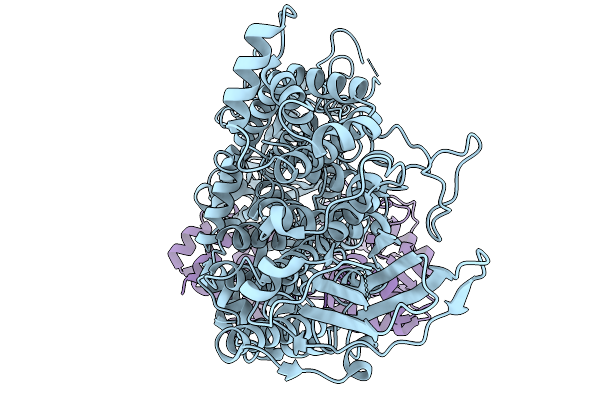 |
Pseudomonas Putida Pore-Forming Toxin Tke5 In Complex With Its Cognate Type Vi Adaptor Protein Tap3
Organism: Pseudomonas putida kt2440
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: TOXIN |
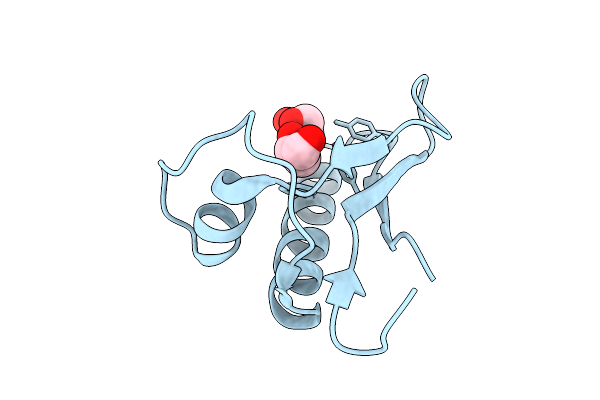 |
The Crystal Structure Of Nuclease Encoded By Gene Paop5_157 From Phage Paop5
Organism: Pseudomonas phage paop5
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: VIRAL PROTEIN Ligands: CL, GOL |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: DNA BINDING PROTEIN Ligands: ACY, SO4, HCI |
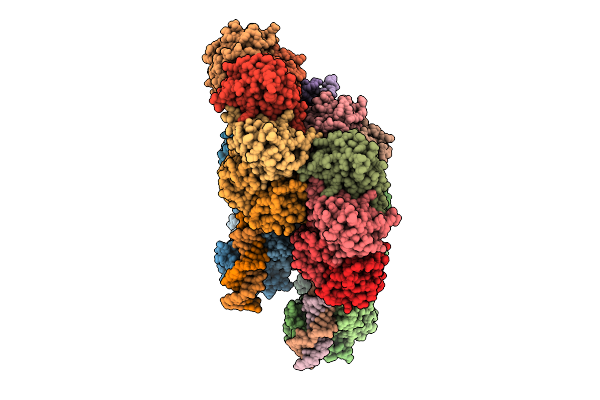 |
Crystal Structure Of The Pseudomonas Putida Xre-Res Toxin-Antitoxin Complex Bound To Promoter Dna
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: GENE REGULATION |
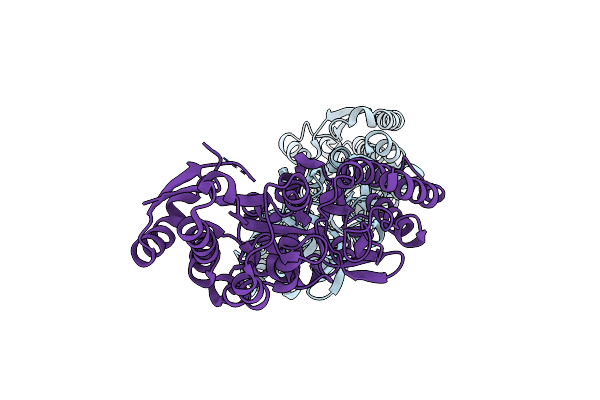 |
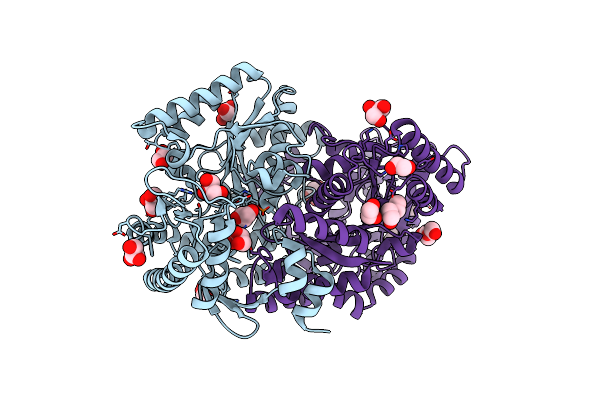
Crystal Structure Of Maltodextrin-Binding Protein Sps0871 From Streptococcus Pyogenes At 2.20 A
Organism: Streptococcus pyogenes m1 gas
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: SUGAR BINDING PROTEIN |
 |
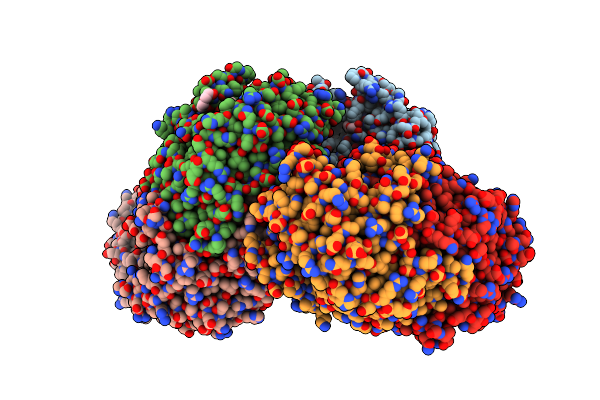
Crystal Structure Of Omega Transaminase Ta_2799 From Pseudomonas Putida Kt2440
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: TRANSFERASE Ligands: GOL, EDO, PEG |
 |
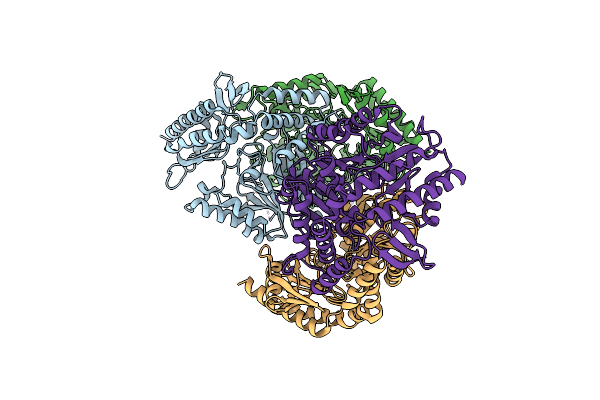
Crystal Structure Of The L322F Mutant Of Omega Transaminase Ta_2799 From Pseudomonas Putida Kt2440
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: TRANSFERASE Ligands: PEG, EDO, SO4, GOL |
 |
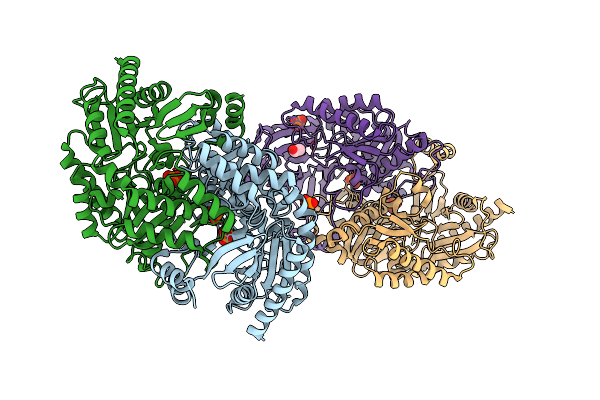
Crystal Structure Of The Open State Of Omega Transaminase Ta_5182 From Pseudomonas Putida Kt2440
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: TRANSFERASE Ligands: SO4 |
 |
Crystal Structure Of The Closed State Of The Omega Transaminase Ta_5182 From Pseudomonas Putida Kt2440
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: TRANSFERASE Ligands: PO4, EDO |
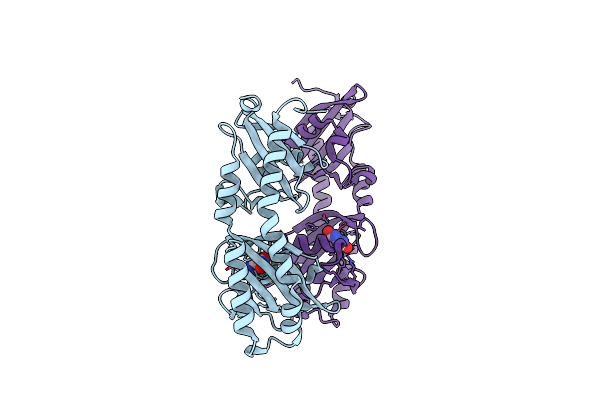 |
Ligand Binding Domain Of The P. Putida Receptor Mcph In Complex With Uric Acid
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-07-24 Classification: SIGNALING PROTEIN Ligands: URC |
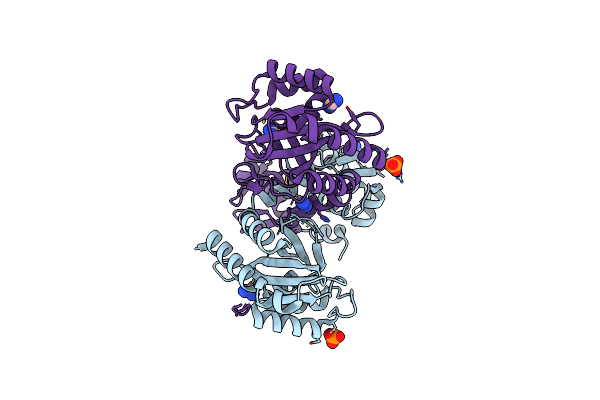 |
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Release Date: 2024-03-20 Classification: TRANSCRIPTION Ligands: SEY, PO4, CL |
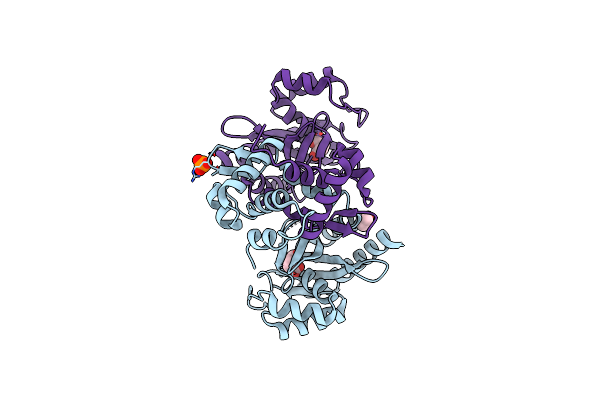 |
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Release Date: 2024-03-20 Classification: TRANSCRIPTION Ligands: SIN, EDO, PO4, CL |
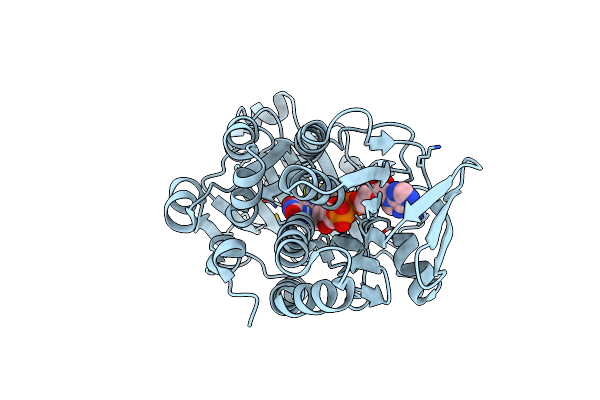 |
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2024-02-21 Classification: OXIDOREDUCTASE Ligands: FAD, PYS |
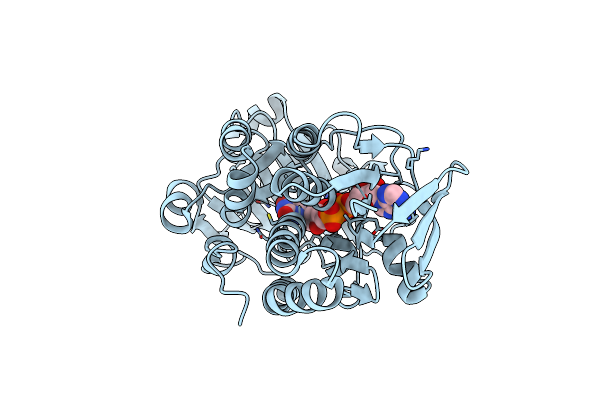 |
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2024-02-21 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Crystal Structure Of A Chimeric Lov-Histidine Kinase Sb2F1 (Asymmetrical Variant, Trigonal Form With Long C Axis)
Organism: Pseudomonas putida kt2440, Bradyrhizobium diazoefficiens usda 110
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2024-01-10 Classification: SIGNALING PROTEIN Ligands: ATP, FMN |
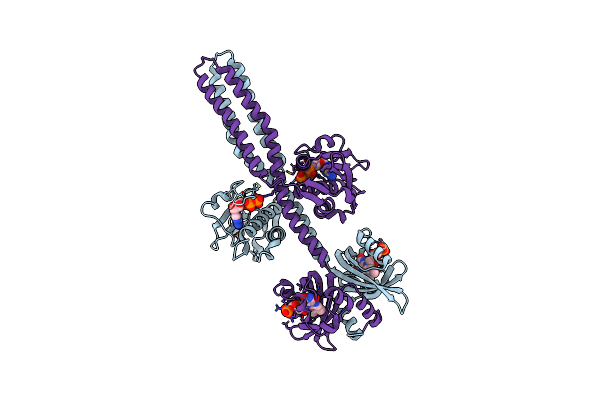 |
Crystal Structure Of A Chimeric Lov-Histidine Kinase Sb2F1-I66R Mutant (Asymmetrical Variant, Trigonal Form With Long C Axis)
Organism: Pseudomonas putida kt2440, Bradyrhizobium diazoefficiens (strain jcm 10833 / bcrc 13528 / iam 13628 / nbrc 14792 / usda 110)
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2024-01-10 Classification: SIGNALING PROTEIN Ligands: ATP, FMN |
 |
Crystal Structure Of A Chimeric Lov-Histidine Kinase Sb2F1-I66R Mutant (Light State; Asymmetrical Variant, Trigonal Form With Long C Axis)
Organism: Pseudomonas putida kt2440, Bradyrhizobium diazoefficiens (strain jcm 10833 / bcrc 13528 / iam 13628 / nbrc 14792 / usda 110)
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2024-01-10 Classification: SIGNALING PROTEIN Ligands: ATP, JGC |
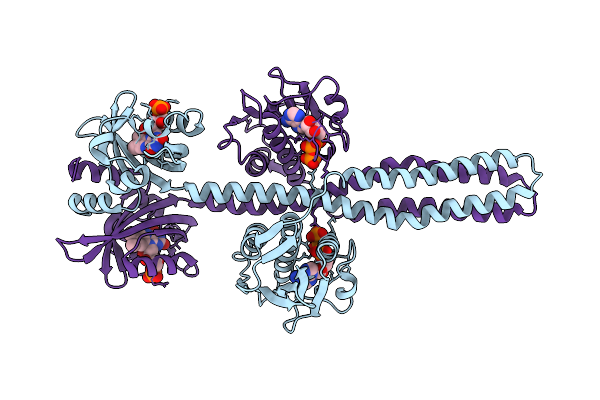 |
Crystal Structure Of A Chimeric Lov-Histidine Kinase Sb2F1 (Asymmetrical Variant, Trigonal Form With Long C-Axis)
Organism: Pseudomonas putida kt2440, Bradyrhizobium diazoefficiens usda 110
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2023-12-27 Classification: SIGNALING PROTEIN Ligands: ATP, FMN |
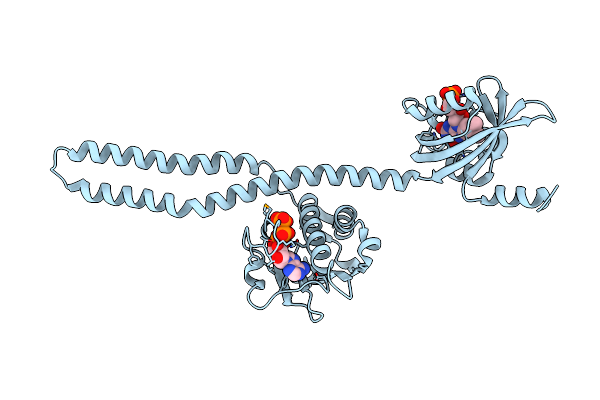 |
Crystal Structure Of A Chimeric Lov-Histidine Kinase Sb2F1 (Symmetrical Variant, Trigonal Form With Short C-Axis)
Organism: Pseudomonas putida kt2440, Bradyrhizobium diazoefficiens usda 110
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2023-12-20 Classification: SIGNALING PROTEIN Ligands: ATP, FMN |
 |
Crystal Structure Of Ppsb1-Lov Protein From Pseudomonas Putida With Covalent Fmn
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-11-29 Classification: SIGNALING PROTEIN Ligands: FMN, NI |