Search Count: 5,729
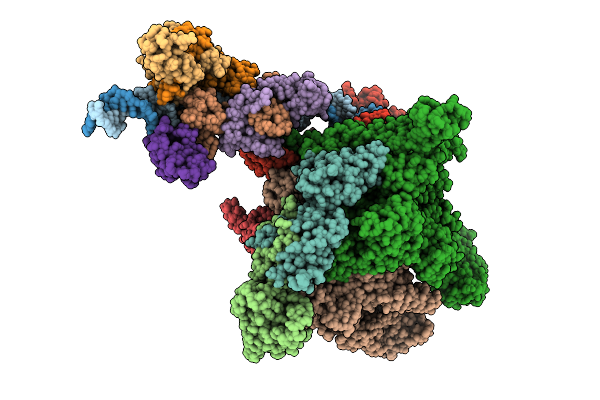 |
Cryo-Em Structure Of E.Coli Transcription Initiation Complex With Escherichia Phage Mu Late Transcription Activator C
Organism: Escherichia coli (strain k12), Escherichia phage mu
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION/DNA |
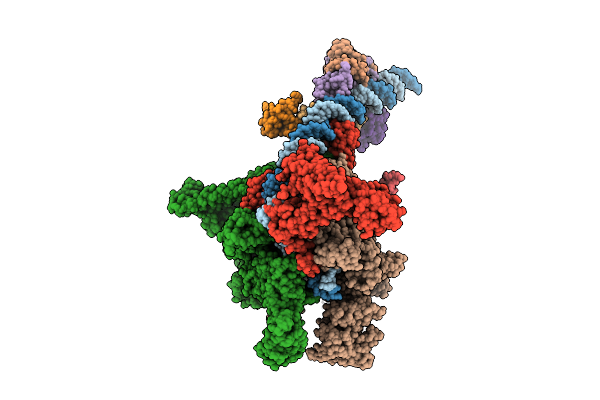 |
Cryo-Em Structure Of E.Coli Transcription Initiation Complex With Escherichia Phage Mu Middle Transcription Activator Mor
Organism: Escherichia coli (strain k12), Escherichia phage mu
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION/DNA |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: DNA BINDING PROTEIN Ligands: ACY, SO4, HCI |
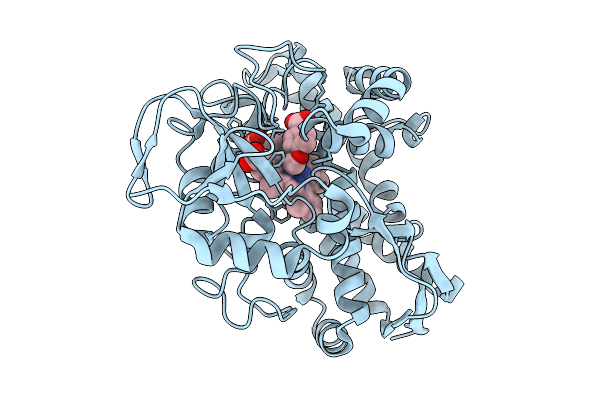 |
The High-Resolution Crystal Structure Of Wt Cyp199A4 Bound To 4-Methoxybenzoic Acid At 100 K
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: OXIDOREDUCTASE Ligands: HEM, ANN, CL |
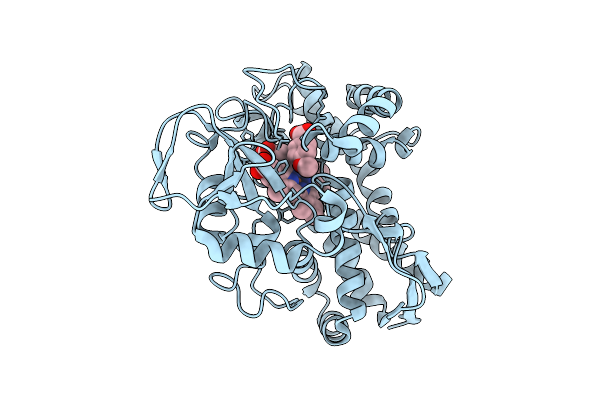 |
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: HEM, ANN, CL |
 |
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: MW7, HEM, CL, ACT |
 |
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: HEM, 5R0, CL, MG |
 |
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: HEM, 5R0, CL |
 |
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: HEM, 4MI |
 |
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: ANN, HEM, CL |
 |
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: HEM, EGM, CL |
 |
The Crystal Structure Of F182Aqe Cyp199A4 Bound To 4-Methylthiobenzoic Acid
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: 4MI, HEM |
 |
Crystal Structure Of The Pseudomonas Putida Xre-Res Toxin-Antitoxin Complex Bound To Promoter Dna
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: GENE REGULATION |
 |
The Crystal Structure Of Wild-Type Cyp199A4 Bound To 4-Trifluoromethoxybenzoic Acid
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: OXIDOREDUCTASE Ligands: M4F, HEM, CL |
 |
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: OXIDOREDUCTASE Ligands: PHB, HEM, CL |
 |
The Crystal Structure Of Wild-Type Cyp199A4 Bound To 4-(Hydroxymethyl)Benzoic Acid
Organism: Rhodopseudomonas palustris haa2
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: OXIDOREDUCTASE Ligands: E5X, HEM, CL, MG |
 |
Crystal Structure Of Maltodextrin-Binding Protein Sps0871 From Streptococcus Pyogenes At 2.20 A
Organism: Streptococcus pyogenes m1 gas
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: SUGAR BINDING PROTEIN |
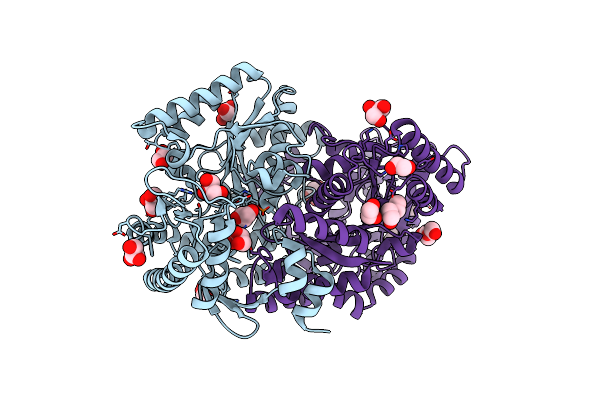 |
Crystal Structure Of Omega Transaminase Ta_2799 From Pseudomonas Putida Kt2440
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: TRANSFERASE Ligands: GOL, EDO, PEG |
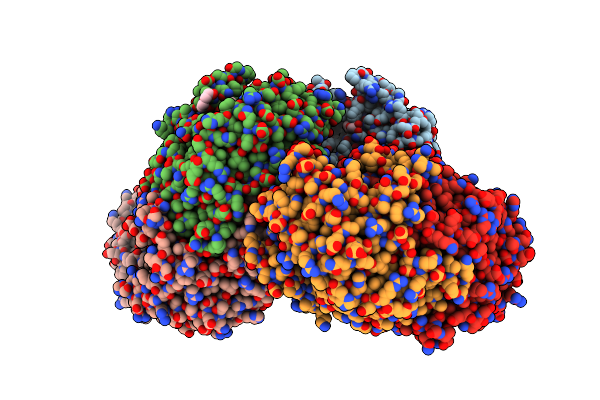 |
Crystal Structure Of The L322F Mutant Of Omega Transaminase Ta_2799 From Pseudomonas Putida Kt2440
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: TRANSFERASE Ligands: PEG, EDO, SO4, GOL |
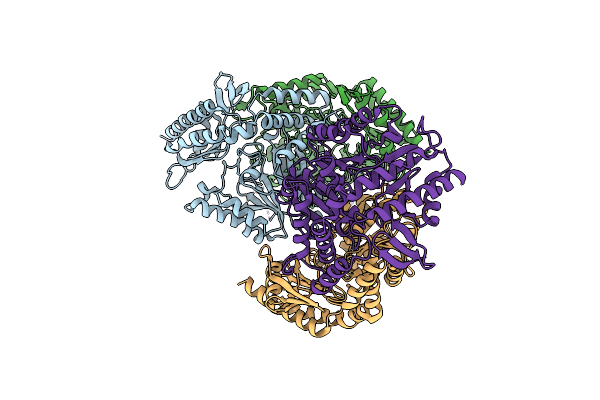 |
Crystal Structure Of The Open State Of Omega Transaminase Ta_5182 From Pseudomonas Putida Kt2440
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: TRANSFERASE Ligands: SO4 |

