Search Count: 130
 |
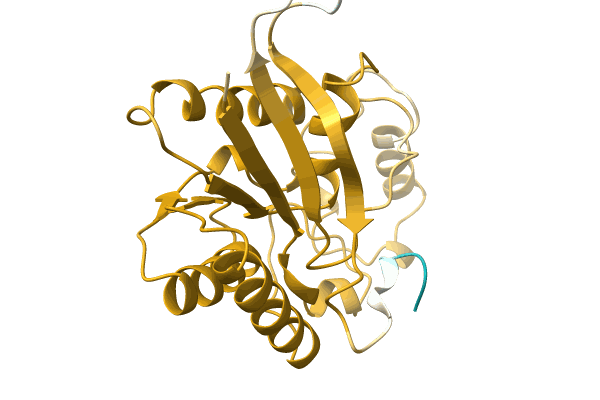
Crystal Structure Of Steroid Aldehyde Dehydrogenase (Sad) From Caldimonas Tepidiphilia
Organism: Caldimonas tepidiphila
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: MPD, TRS |
 |
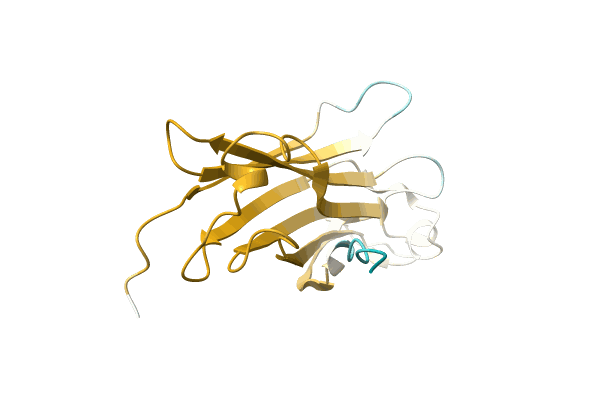
Organism: Hahella
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2023-10-11 Classification: OXIDOREDUCTASE |
 |
Organism: Podospora anserina (strain s / atcc mya-4624 / dsm 980 / fgsc 10383)
Method: SOLID-STATE NMR Release Date: 2018-10-10 Classification: PROTEIN FIBRIL |
 |
Organism: Lactobacillus delbrueckii
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2018-07-25 Classification: TRANSPORT PROTEIN |
 |
Organism: Lactobacillus delbrueckii
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2016-04-06 Classification: TRANSPORT PROTEIN Ligands: ANP |
 |
Crystal Structure Of Laglidadg Meganuclease I-Panmi With Coordinated Calcium Ions
Organism: Podospora anserina, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-03-30 Classification: HYDROLASE/DNA Ligands: CA, GOL |
 |
Crystal Structure Of The Ctp1L Endolysin Reveals How Its Activity Is Regulated By A Secondary Translation Product
Organism: Clostridium phage phictp1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-12-30 Classification: STRUCTURAL PROTEIN Ligands: SO4, GOL, 1PE |
 |
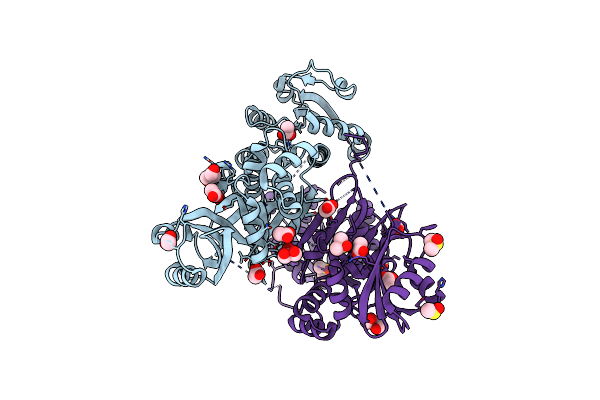
Organism: Staphylococcus aureus m1064
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-05-06 Classification: OXIDOREDUCTASE Ligands: ACT, GOL, PEG, DMS |
 |
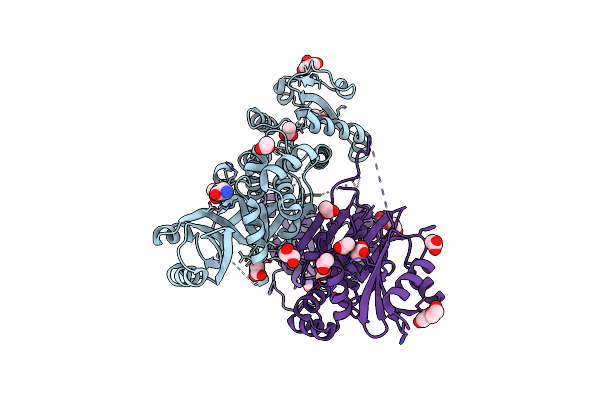
Organism: Staphylococcus aureus m1064
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-05-06 Classification: OXIDOREDUCTASE Ligands: ACT, GOL, PEG, DMS |
 |
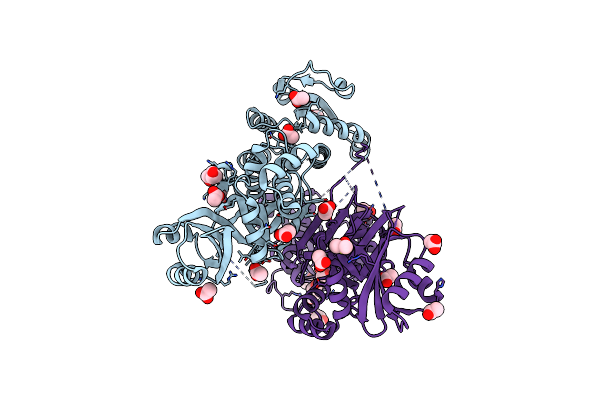
Organism: Staphylococcus aureus m1064
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-05-06 Classification: OXIDOREDUCTASE Ligands: ACT, GOL, LYS, PEG |
 |
Organism: Staphylococcus aureus m1064
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-05-06 Classification: OXIDOREDUCTASE Ligands: ACT, PEG, GOL, DMS |
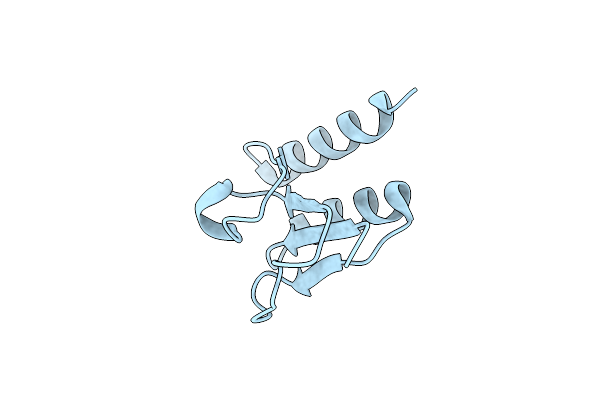 |
C-Terminal Domain Of Ctp1L Endolysin Mutant V195P That Reduces Autoproteolysis
Organism: Clostridium phage phictp1
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2014-08-06 Classification: HYDROLASE |
 |
Organism: Podospora anserina
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2013-04-17 Classification: HYDROLASE Ligands: CA, HG, TLA |
 |
Crystal Structure Of Putative Phosphoglycolate Phosphatase (Yp_619066.1) From Lactobacillus Delbrueckii Subsp. Bulgaricus Atcc Baa-365 At 1.51 A Resolution
Organism: Lactobacillus delbrueckii
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2006-08-15 Classification: HYDROLASE |
 |
 |
 |
 |
 |
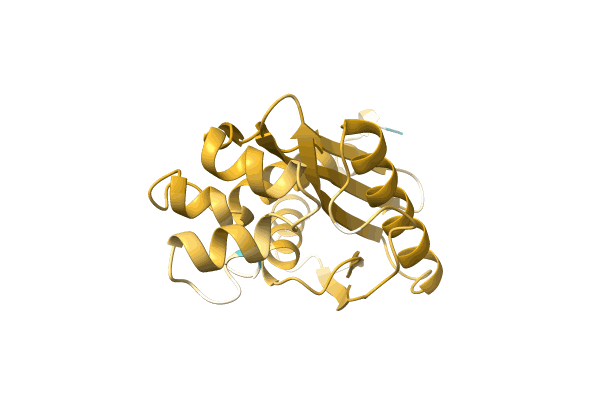 |

