Search Count: 72
 |
Crystal Structure Of C1 Domain From Surface Protein Slpm Of Lactobacillus Brevis
Organism: Levilactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-06-11 Classification: STRUCTURAL PROTEIN |
 |
Organism: Levilactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: NAP, EDO, PEG, PGE, ACY, MG |
 |
Organism: Levilactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: NAP, SO4, MG |
 |
Organism: Levilactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: NAP, PG4, MG, EDO |
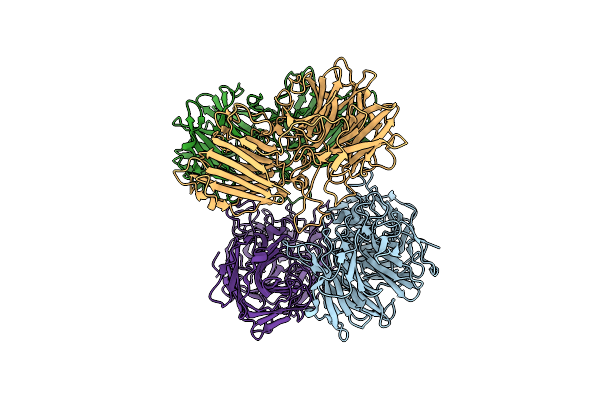 |
The Molecular Structure Of A Beta-1,4-D-Xylosidase From The Probiotic Bacterium Levilactobacillus Brevis
Organism: Levilactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-01-15 Classification: CARBOHYDRATE |
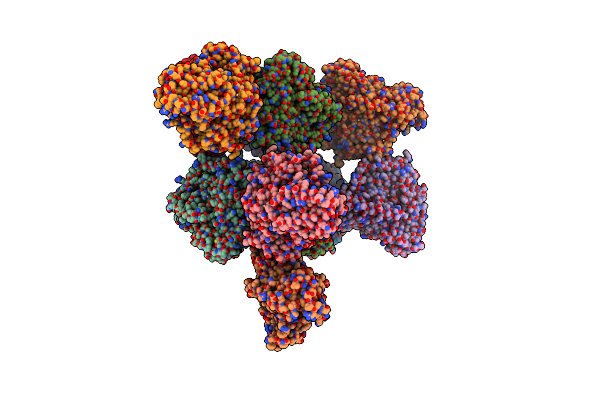 |
Organism: Enhygromyxa salina
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-11-27 Classification: OXIDOREDUCTASE Ligands: CD |
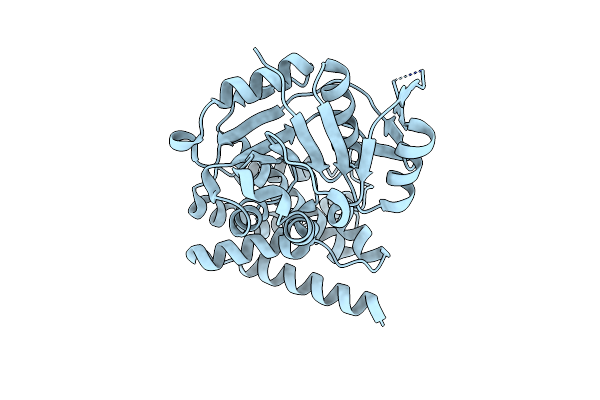 |
The Crystal Structure Of D-Mandelate Dehydrogenase From Lactobacillus Brevis
Organism: Levilactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-10-02 Classification: OXIDOREDUCTASE |
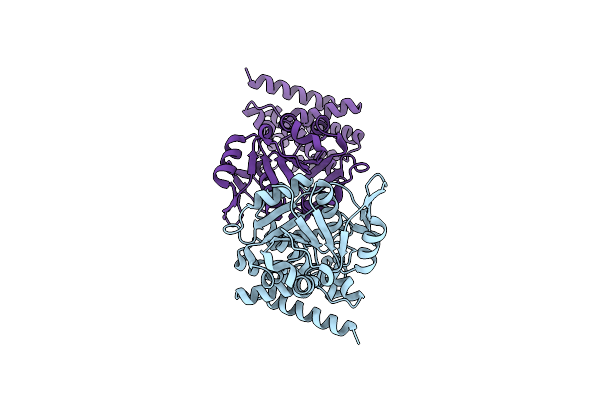 |
Organism: Levilactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-10-02 Classification: OXIDOREDUCTASE |
 |
Organism: Levilactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2024-10-02 Classification: OXIDOREDUCTASE |
 |
Organism: Enhygromyxa salina
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2024-02-07 Classification: LUMINESCENT PROTEIN Ligands: SO4 |
 |
Organism: Enhygromyxa salina
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2023-05-24 Classification: LIPID BINDING PROTEIN Ligands: PLM |
 |
Organism: Enhygromyxa salina
Method: X-RAY DIFFRACTION Resolution:1.03 Å Release Date: 2023-05-24 Classification: LIPID BINDING PROTEIN |
 |
Organism: Lactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-10-28 Classification: OXIDOREDUCTASE Ligands: MG |
 |
Organism: Lactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2020-02-19 Classification: OXIDOREDUCTASE Ligands: MG |
 |
Organism: Lactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2020-02-19 Classification: OXIDOREDUCTASE Ligands: EDO, PEG, PGE, MG |
 |
Organism: Lactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2020-02-19 Classification: OXIDOREDUCTASE Ligands: MG, EDO |
 |
X-Ray Structure Of Lactobacillus Brevis Alcohol Dehydrogenase Mutant T102E_Q126K
Organism: Lactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-02-19 Classification: OXIDOREDUCTASE Ligands: EDO, MG |
 |
X-Ray Structure Of Lactobacillus Brevis Alcohol Dehydrogenase Mutant K32A_Q126K
Organism: Lactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-02-19 Classification: OXIDOREDUCTASE Ligands: EDO, MG, PEG |
 |
Organism: Lactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2020-02-19 Classification: OXIDOREDUCTASE Ligands: PEG, PG4, EDO, MG, CL, PGE |
 |
Organism: Chromohalobacter salexigens dsm 3043
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2019-09-11 Classification: OXIDOREDUCTASE Ligands: DTT, PO4 |

