Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
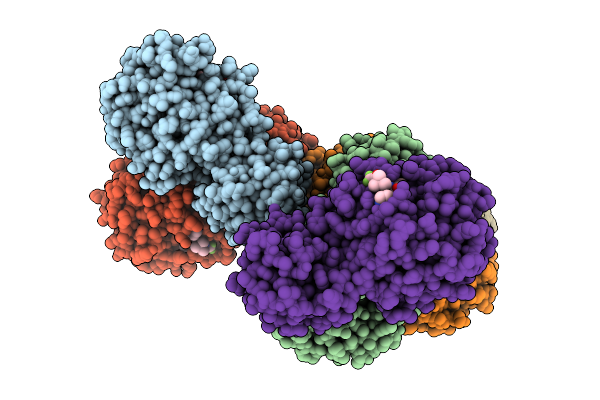 |
Crystal Structure Of Pdcov 3Cl Protease (3Clpro) In Complex With Compound 8
Organism: Porcine deltacoronavirus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: A1EPZ |
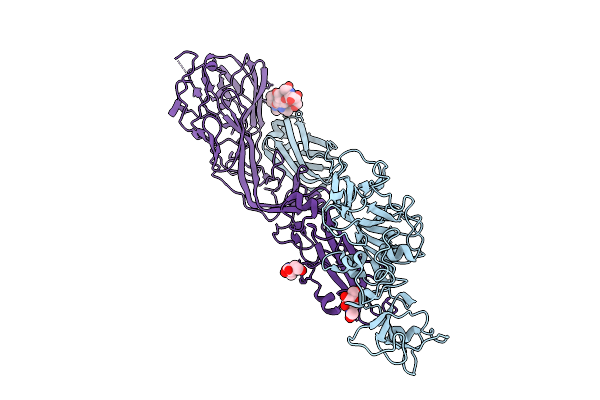 |
Organism: Rift valley fever virus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: NAG |
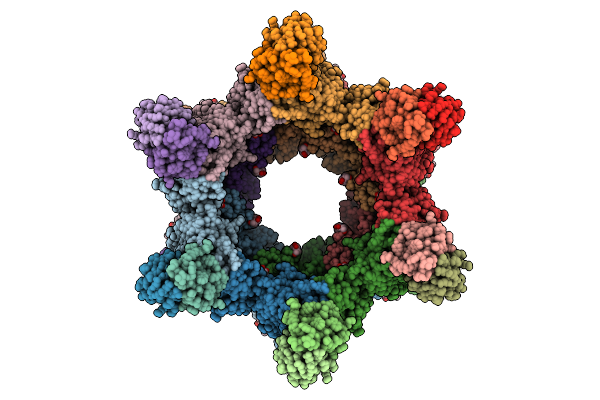 |
Organism: Rift valley fever virus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
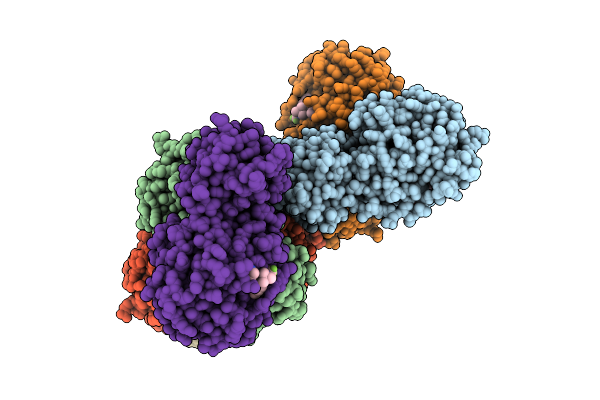 |
Crystal Structure Of Pdcov 3Cl Protease (3Clpro) In Complex With Nirmatrelvir
Organism: Porcine deltacoronavirus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: 4WI |
 |
Crystal Structure Of Pdcov 3Cl Protease (3Clpro) In Complex With Compound 1
Organism: Porcine deltacoronavirus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: A1D7G |
 |
Crystal Structure Of Pdcov 3Cl Protease (3Clpro) In Complex With Compound 3
Organism: Porcine deltacoronavirus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: A1D7N |
 |
Crystal Structure Of Pdcov 3Cl Protease (3Clpro) In Complex With Compound 6
Organism: Porcine deltacoronavirus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: A1D7M |
 |
Organism: Homo sapiens, Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
 |
Organism: Rift valley fever virus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN Ligands: GOL, PEG |
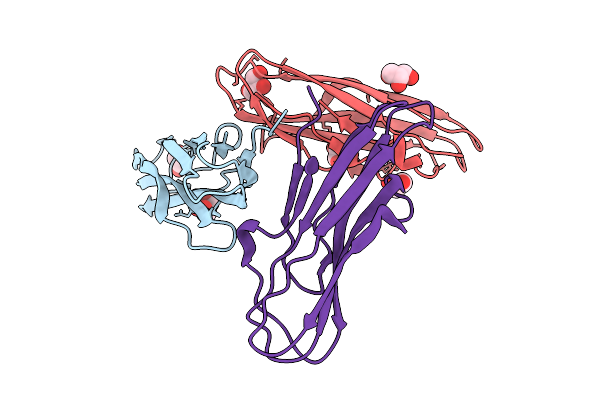 |
Organism: Pseudanabaena phage pan3
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRUS Ligands: IOD, GOL |
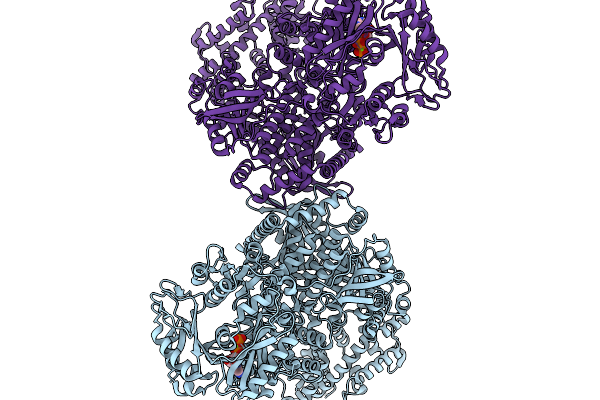 |
Cryo-Em Structure Of Acetyl-Coa Carboxyltransferase Holoenzyme Dimer From Shewanella Oneidensis, Mutant E1238A, Complexed With Adp
Organism: Shewanella oneidensis mr-1
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: LIGASE Ligands: ADP, MG |
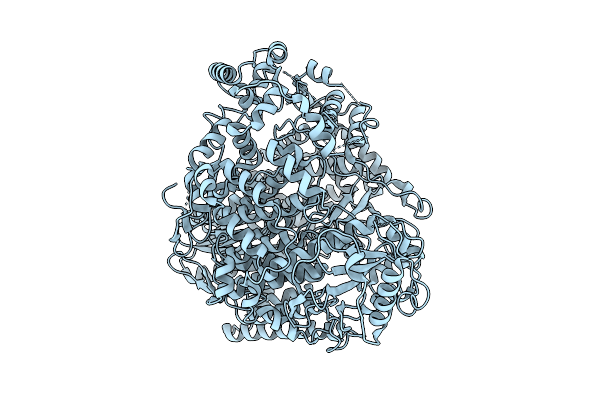 |
Organism: Rift valley fever virus (strain zh-548 m12)
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: VIRAL PROTEIN |
 |
Organism: Staphylococcus aureus subsp. aureus n315
Method: SOLUTION NMR Release Date: 2025-10-15 Classification: DNA BINDING PROTEIN |
 |
Organism: Staphylococcus aureus subsp. aureus n315, Synthetic construct
Method: SOLUTION NMR Release Date: 2025-10-15 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of Steroid Aldehyde Dehydrogenase (Sad) From Caldimonas Tepidiphilia
Organism: Caldimonas tepidiphila
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: MPD, TRS |
 |
Organism: Clostridium phage phicd111
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: ANTIBIOTIC Ligands: ZN |
 |
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: LIGASE Ligands: LYS, GOL |
 |
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: LYASE Ligands: SO4 |
 |
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: LYASE Ligands: PEG |
 |
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: LYASE Ligands: PGU |