Search Count: 207
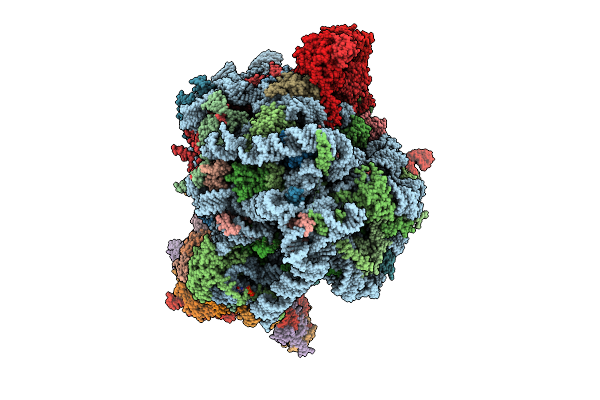 |
Human Quaternary Complex Of A Translating 80S Ribosome, Nac, Metap1 And Natd
Organism: Homo sapiens, Saccharomyces cerevisiae s288c, Aequorea victoria, Brachypodium distachyon
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: ZN, COA, GTP |
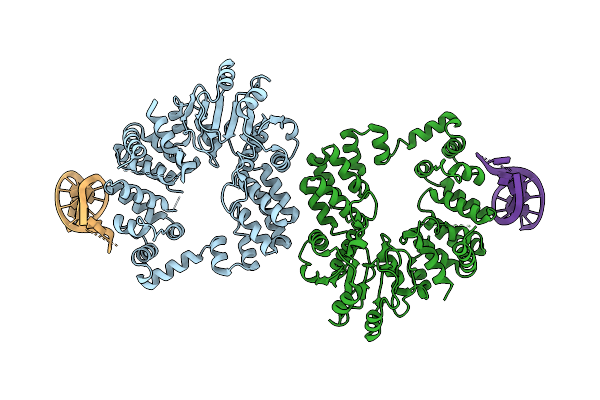 |
Organism: Thermus phage philo
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: RNA BINDING PROTEIN/RNA |
 |
Organism: Thermus phage philo
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: RNA BINDING PROTEIN Ligands: CA |
 |
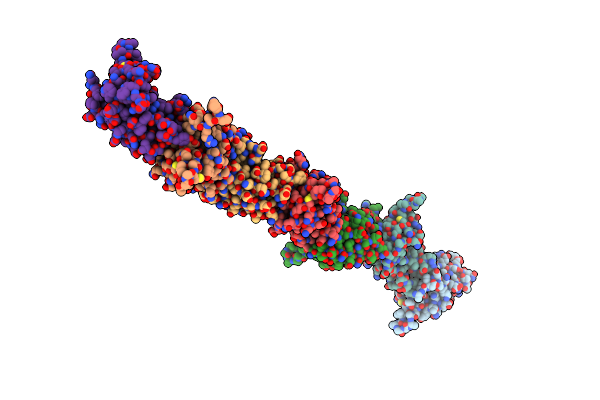
Organism: Brachypodium distachyon
Method: ELECTRON MICROSCOPY Release Date: 2021-05-19 Classification: MEMBRANE PROTEIN Ligands: PLC, SPH |
 |
Organism: Moniliophthora perniciosa
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2017-08-30 Classification: LIPID BINDING PROTEIN |
 |
Organism: Moniliophthora perniciosa
Method: X-RAY DIFFRACTION Resolution:2.92 Å Release Date: 2017-08-30 Classification: LIPID BINDING PROTEIN Ligands: SEY |
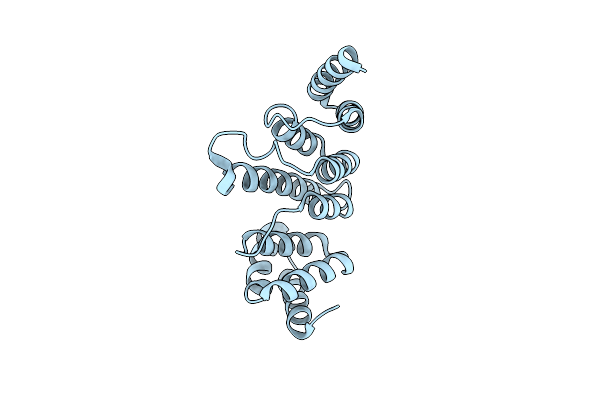 |
Organism: Brachypodium distachyon
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-10-30 Classification: RNA BINDING PROTEIN |
 |
Organism: Brachypodium distachyon
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2013-10-30 Classification: SPLICING/RNA |
 |
Organism: Brachypodium distachyon
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2013-10-30 Classification: SPLICING/RNA |
 |
Organism: Moniliophthora perniciosa
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2012-07-11 Classification: UNKNOWN FUNCTION Ligands: ACT, ZN, CL, NA |
 |
Organism: Moniliophthora perniciosa
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2012-07-11 Classification: UNKNOWN FUNCTION |
 |
Organism: Moniliophthora perniciosa
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2012-07-11 Classification: UNKNOWN FUNCTION |
 |
Organism: Moniliophthora perniciosa
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2012-07-11 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Cyclophilin A From Moniliophthora Perniciosa In Complex With Cyclosporin A
Organism: Moniliophthora perniciosa, Tolypocladium inflatum
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2011-11-23 Classification: ISOMERASE/IMMUNOSUPPRESSANT |
 |
Crystal Structure Of Necrosis And Ethylene Inducing Protein 2 From The Causal Agent Of Cocoa'S Witches Broom Disease
Organism: Moniliophthora perniciosa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-11-02 Classification: TOXIN Ligands: ZN, NA |
 |
Organism: Moniliophthora perniciosa
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2011-08-10 Classification: ISOMERASE |
 |
Organism: Moniliophthora perniciosa
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2009-11-17 Classification: LIPID BINDING PROTEIN Ligands: ZN, MES |
 |
Organism: Clostridium limosum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-04-01 Classification: TRANSFERASE Ligands: SO4 |
 |
 |

