Search Count: 55
 |
Crystal Structure Of Ctag From Ruminiclostridium Cellulolyticum (P2(1)-Small)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: LIGASE Ligands: SO4, CO3 |
 |
Crystal Structure Of Ctag From Ruminiclostridium Cellulolyticum (P2(1)-Medium)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: LIGASE Ligands: GOL |
 |
Crystal Structure Of The Ctag_C11A Variant From Ruminiclostridium Cellulolyticum (P2(1)-Small)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: LIGASE |
 |
Crystal Structure Of The Ctag_C11A Variant From Ruminiclostridium Cellulolyticum (P2(1)2(1)2(1)-Small)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: LIGASE Ligands: NI, IMD, TRS |
 |
Crystal Structure Of The Ctag_H128A Variant From Ruminiclostridium Cellulolyticum (P2(1)-Small)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: LIGASE Ligands: SO4, CO3 |
 |
Crystal Structure Of The Ctag_H128A Variant From Ruminiclostridium Cellulolyticum (P2(1)2(1)2(1)-Medium)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: LIGASE |
 |
Crystal Structure Of The Ctag_H128A Variant From Ruminiclostridium Cellulolyticum In Complex With Phba (P2(1)-Medium)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: LIGASE Ligands: HBA, PO4 |
 |
Crystal Structure Of The Ctag_H128A Variant From Ruminiclostridium Cellulolyticum In Complex With Phba (P2(1)2(1)2(1)-Medium)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: LIGASE Ligands: HBA, GOL |
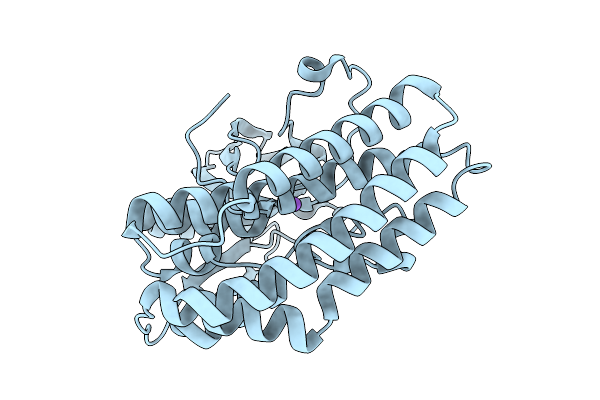 |
Crystal Structure Of The Ctag_D144N Variant From Ruminiclostridium Cellulolyticum (P2(1)-Small)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: LIGASE |
 |
Crystal Structure Of Duf1735 Domain-Containing Protein (Gh18-Like) From Bacteroides Faecium At 2.7 A Resolution (Space Group C2)
Organism: Bacteroides faecium
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2024-05-29 Classification: SUGAR BINDING PROTEIN |
 |
Crystal Structure Of Duf1735-Domain Containing Protein (Gh18-Like) From Bacteroides Faecium At 2.9 A Resolution (Space Group P21)
Organism: Bacteroides faecium
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2024-05-29 Classification: SUGAR BINDING PROTEIN |
 |
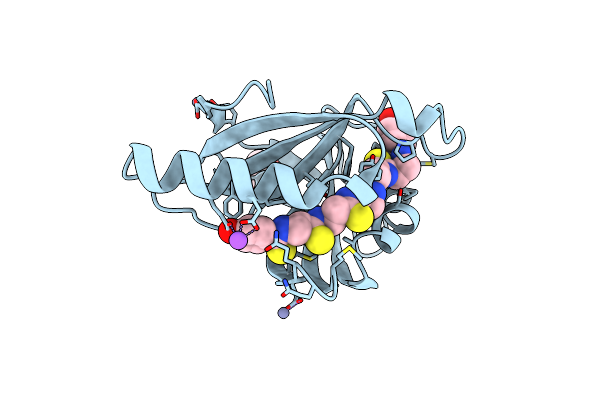
Crystal Structure Of An Alpha/Beta-Hydrolase Enzyme From Chloroflexus Sp. Ms-G (202)
Organism: Chloroflexus sp. ms-g
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2022-12-28 Classification: HYDROLASE |
 |
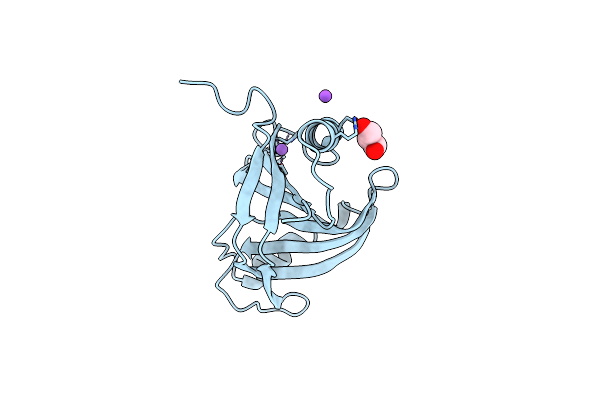
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-11-09 Classification: LIPID BINDING PROTEIN Ligands: IQ4, ZN, NA, TRS |
 |
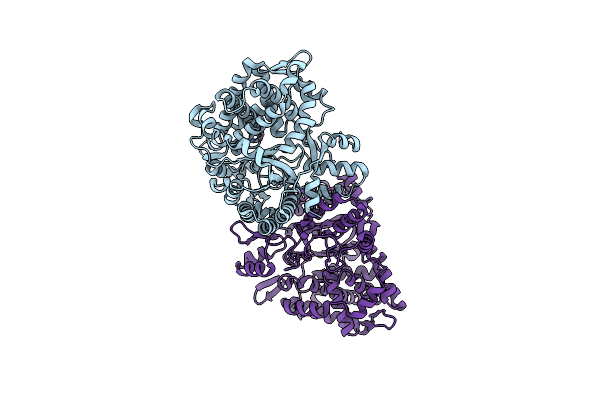
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-11-09 Classification: LIPID BINDING PROTEIN Ligands: GOL, NA |
 |
Organism: Corallococcus sp. egb
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-06-12 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of The Intrinsic Colistin Resistance Enzyme Icr(Mc) From Moraxella Catarrhalis, Catalytic Domain, Thr315Ala Mutant Di-Zinc And Peg Complex
Organism: Moraxella sp. hmsc061h09
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-01-31 Classification: TRANSFERASE Ligands: ZN, 15P, CL |
 |
Crystal Structure Of The Intrinsic Colistin Resistance Enzyme Icr(Mc) From Moraxella Catarrhalis, Catalytic Domain, Thr315Ala Mutant Mono-Zinc And Phosphoethanolamine Complex
Organism: Moraxella sp. hmsc061h09
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2018-01-31 Classification: TRANSFERASE Ligands: ZN, OPE, SO4, 15P |
 |
Crystal Structure Of The Intrinsic Colistin Resistance Enzyme Icr(Mc) From Moraxella Catarrhalis, Catalytic Domain, Phosphate-Bound Complex
Organism: Moraxella sp. hmsc061h09
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2018-01-31 Classification: TRANSFERASE Ligands: SO4, GOL, PO4, ACT |
 |
Crystal Structure Of The Intrinsic Colistin Resistance Enzyme Icr(Mc) From Moraxella Catarrhalis, Catalytic Domain, Mono-Zinc Complex
Organism: Moraxella sp. hmsc061h09
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2018-01-31 Classification: TRANSFERASE Ligands: PO4, ZN, SO4, ACT, GOL |
 |
Crystal Structure Of A Family 6 Carbohydrate-Binding Module From Clostridium Cellulolyticum In Complex With Xylose
Organism: Clostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-10-13 Classification: HYDROLASE Ligands: XYP, NA |

