Search Count: 25,382
 |
Organism: Acetivibrio thermocellus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
Organism: Acetivibrio thermocellus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: ZN, ANP |
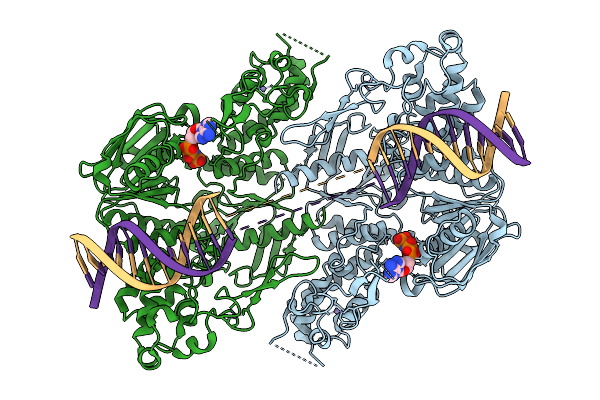 |
C. Thermocellum Uvra In Complex With Unmodified Dna And Amppnp (Basal Conformation)
Organism: Acetivibrio thermocellus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: ZN, ANP |
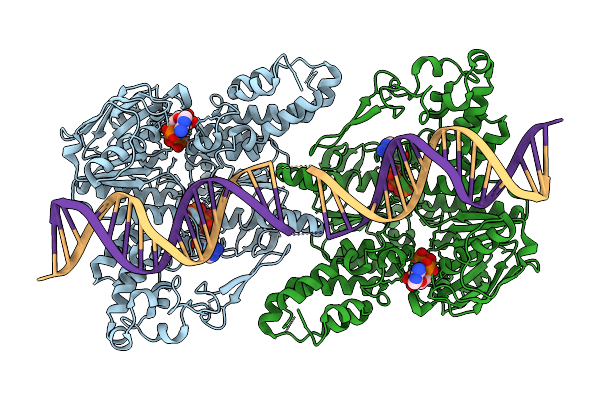 |
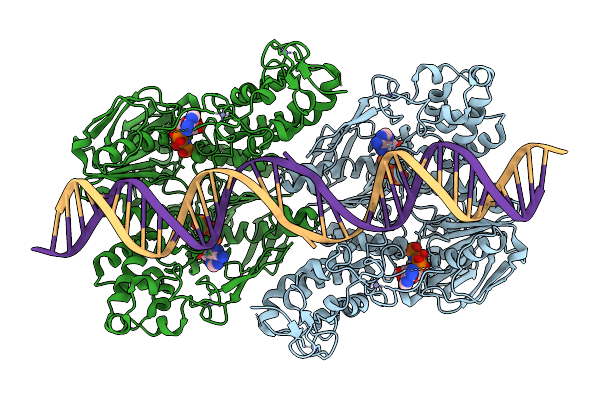
C. Thermocellum Uvra In Complex With Unmodified Dna And Amppnp (Atp-Bound Conformation 1)
Organism: Acetivibrio thermocellus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: ZN, ANP |
 |
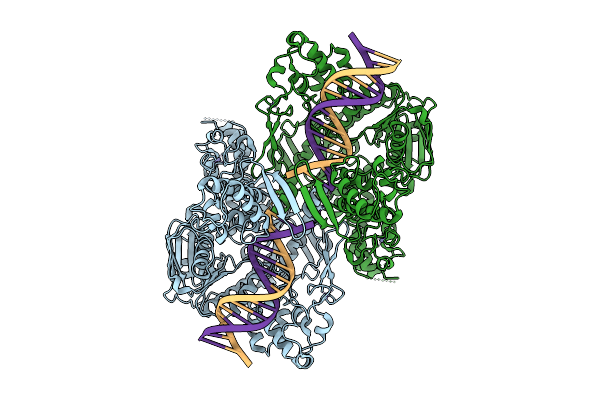
C. Thermocellum Uvra In Complex With Unmodified Dna And Amppnp (Atp-Bound Conformation 2)
Organism: Acetivibrio thermocellus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: ZN, ANP |
 |
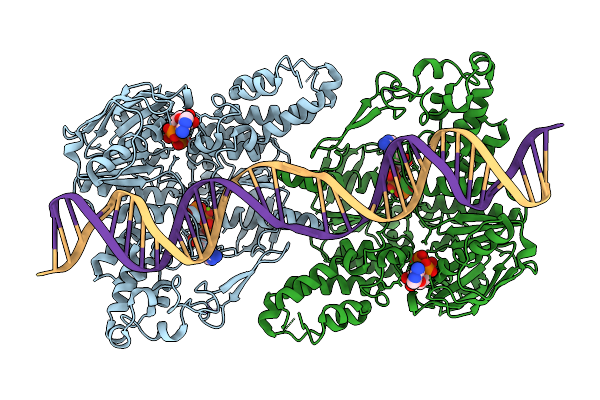
C. Thermocellum Uvra In Complex With Dna With An Abasic Site (Basal Conformation)
Organism: Acetivibrio thermocellus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
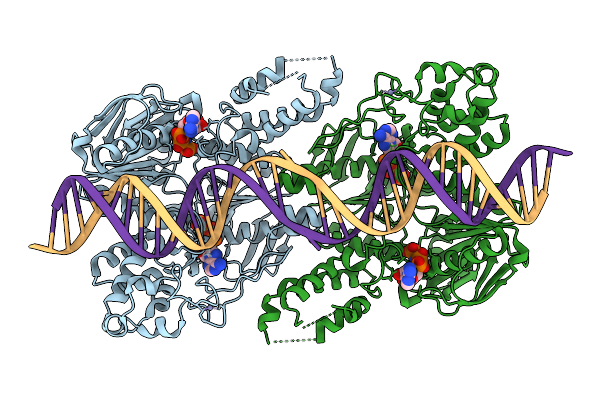
C. Thermocellum Uvra In Complex With Dna With An Abasic Site And Amppnp (Atp-Bound Conformation 1)
Organism: Acetivibrio thermocellus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: ZN, ANP, MG |
 |
C. Thermocellum Uvra In Complex With Dna With An Abasic Site And Amppnp (Atp-Bound Conformation 2)
Organism: Acetivibrio thermocellus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: ZN, ANP |
 |
C. Thermocellum Uvra In Complex With Dna With A Fluorescein Modification (Basal Conformation)
Organism: Acetivibrio thermocellus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
C. Thermocellum Uvra In Complex With Dna With A Fluorescein Modification And Amppnp (Atp-Bound Conformation 1)
Organism: Acetivibrio thermocellus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: ZN, ANP |
 |
C. Thermocellum Uvra (K647A) In Complex With Dna With A Fluorescein Modification (Basal Conformation)
Organism: Acetivibrio thermocellus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
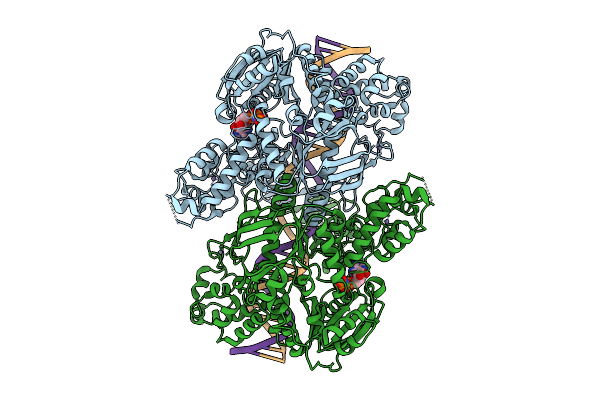
C. Thermocellum Uvra In Complex With Dna With A Fluorescein Modification And Adp (Basal Conformation)
Organism: Acetivibrio thermocellus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: ZN, ADP |
 |
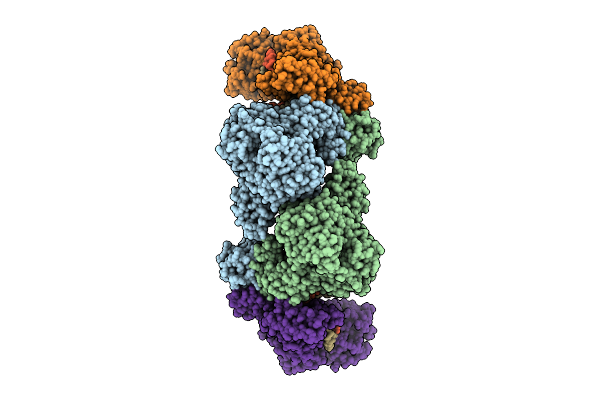
C. Thermocellum Uvra (K39A) In Complex With Dna With A Fluorescein Modification And Amppnp (Basal Conformation)
Organism: Acetivibrio thermocellus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: ZN, ANP |
 |
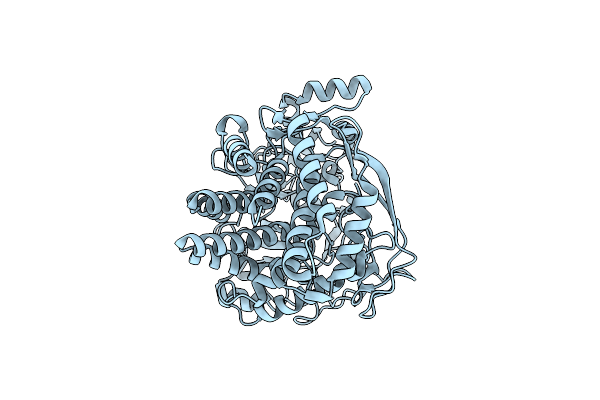
C. Thermocellum Uvra-Uvrb In Complex With Dna With A Fluorescein Modification And Amppnp
Organism: Acetivibrio thermocellus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: ZN, ANP, MG |
 |
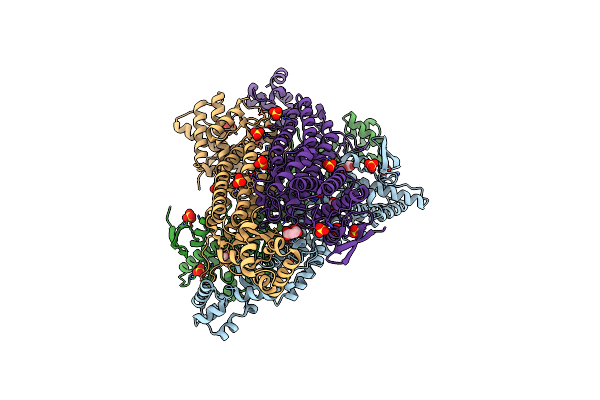
Organism: Acetivibrio thermocellus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: HYDROLASE |
 |
Organism: Staphylococcus aureus subsp. aureus usa300_fpr3757
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-05-14 Classification: HYDROLASE Ligands: FUM, FMT, SO4, CL |
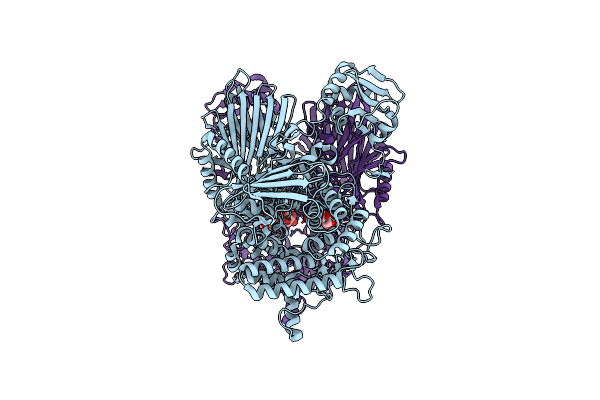 |
Cryo-Em Structure Of Cellodextrin Phosphorylase From Clostridium Thermocellum With Cellodextrin Ligands
Organism: Acetivibrio thermocellus
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: CARBOHYDRATE Ligands: CL |
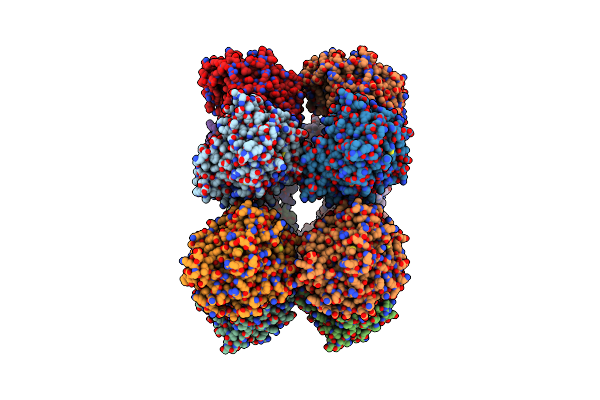 |
Organism: Acetivibrio thermocellus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: TRS |
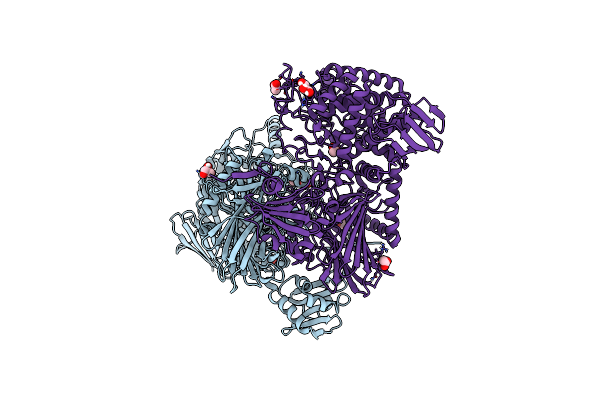 |
Cellodextrin Phosphorylase From Clostridium Thermocellum Mutant - All Cysteine Residues Were Substituted With Serines
Organism: Acetivibrio thermocellus
Method: X-RAY DIFFRACTION Release Date: 2024-12-25 Classification: CARBOHYDRATE Ligands: GOL, PEG, CL, ACT |
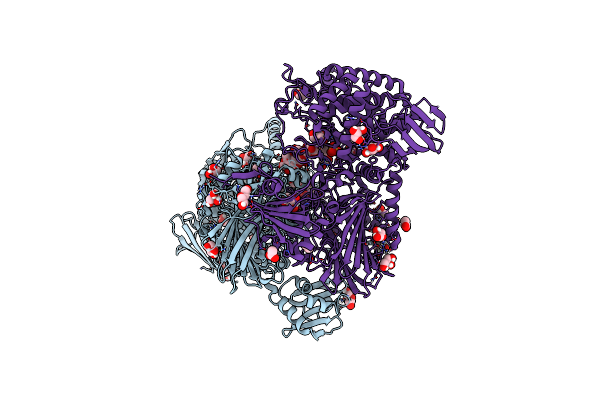 |
Cellodextrin Phosphorylase From Clostridium Thermocellum Mutant - All Cysteine Residues Were Substituted With Serines
Organism: Acetivibrio thermocellus
Method: X-RAY DIFFRACTION Release Date: 2024-12-25 Classification: CARBOHYDRATE Ligands: ACT, TRS, SO4, PEG, GOL, CL |

