Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
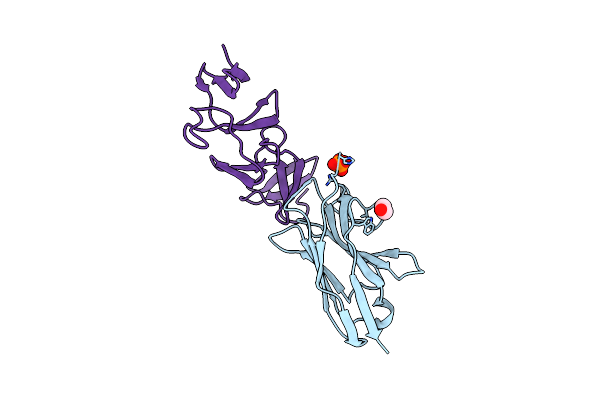 |
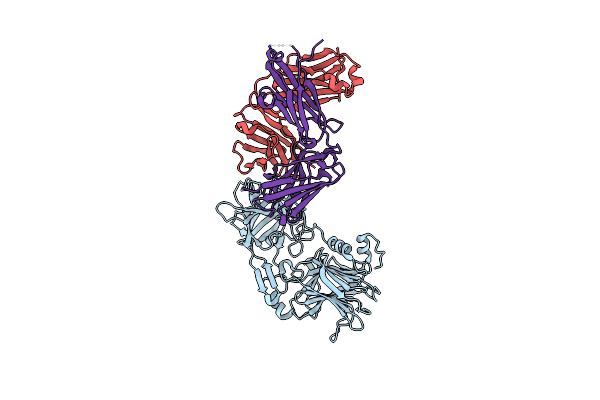
Crystal Structure Of The C. Difficile Toxin A Crops Domain Fragment 2592-2710 Bound To H5.2 Nanobody
Organism: Camelus dromedarius, Clostridioides difficile 342
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-12-18 Classification: TOXIN/IMMUNE SYSTEM Ligands: EDO, PO4 |
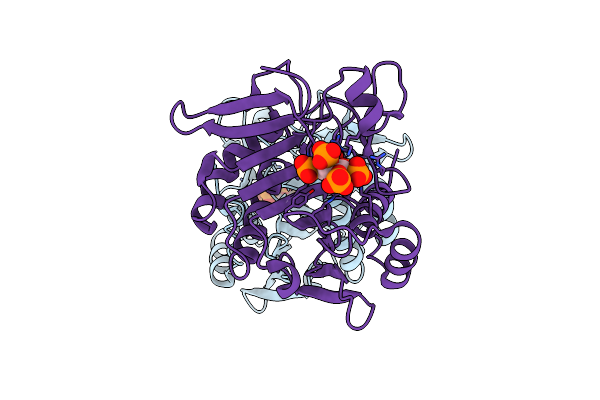 |
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-07-03 Classification: TOXIN Ligands: IHP |
 |
Cryo-Em Structure Of Spike Glycoprotein From Civet Coronavirus Sz3 In Closed Conformation
Organism: Civet sars cov sz3/2003
Method: ELECTRON MICROSCOPY Release Date: 2024-05-15 Classification: VIRAL PROTEIN Ligands: NAG, EIC |
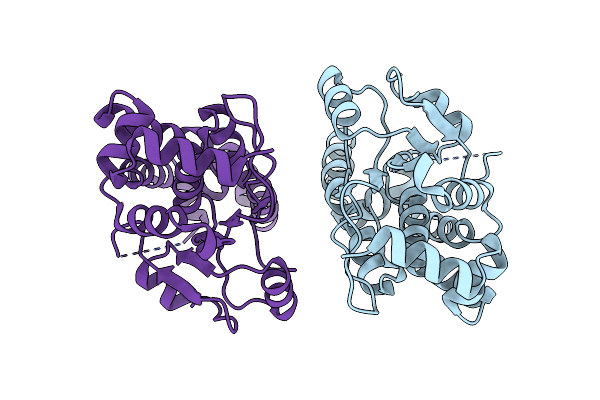 |
Crystal Structure Of Clostridioides Difficile Protein Tyrosine Phosphatase At Ph 8.5
Organism: Clostridioides difficile 6050
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2023-07-05 Classification: HYDROLASE |
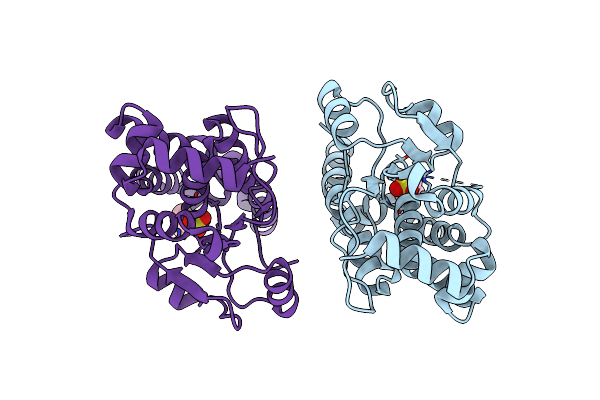 |
Crystal Structure Of Clostridioides Difficile Protein Tyrosine Phosphatase At Ph 7.5
Organism: Clostridioides difficile 6050
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2023-07-05 Classification: HYDROLASE Ligands: EPE, CL |
 |
Organism: Lloviu cuevavirus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: VIRAL PROTEIN |
 |
Organism: Lloviu cuevavirus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: VIRAL PROTEIN |
 |
Apo-Structure Of Serine Hydroxymethyltransferase (Pbzb) Involved In Benzobactin Biosynthesis In P. Chlororaphis Subsp. Piscium Dsm 21509
Organism: Pseudomonas chlororaphis subsp. piscium
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2022-10-12 Classification: BIOSYNTHETIC PROTEIN |
 |
Fnip Family Proteins From Cafeteria Roenbergensis Virus (Crov): Leucine-Rich Repeats With Novel Structural Features
Organism: Cafeteria roenbergensis virus
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2022-08-24 Classification: UNKNOWN FUNCTION |
 |
Structural Insight Into The Interactions Between Lloviu Virus Vp30 And Nucleoprotein
Organism: Lloviu cuevavirus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-06-29 Classification: VIRUS |
 |
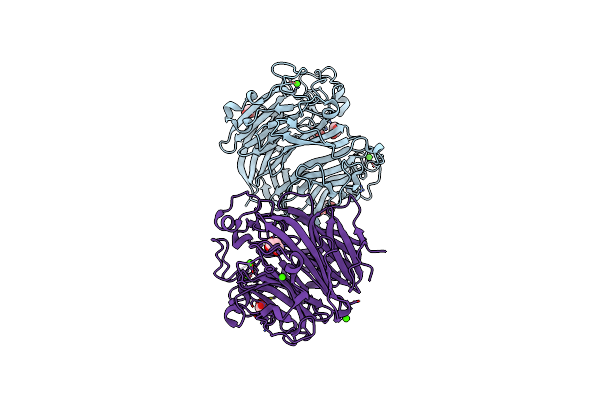
Organism: Clostridium tetani, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2021-04-07 Classification: HYDROLASE |
 |
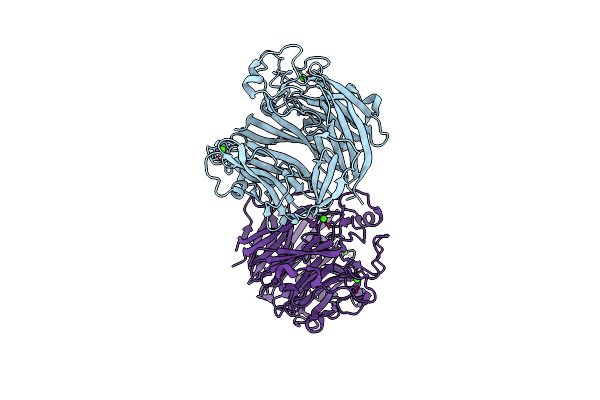
Organism: Catenovulum maritimum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-04-07 Classification: LYASE Ligands: CA, GOL |
 |
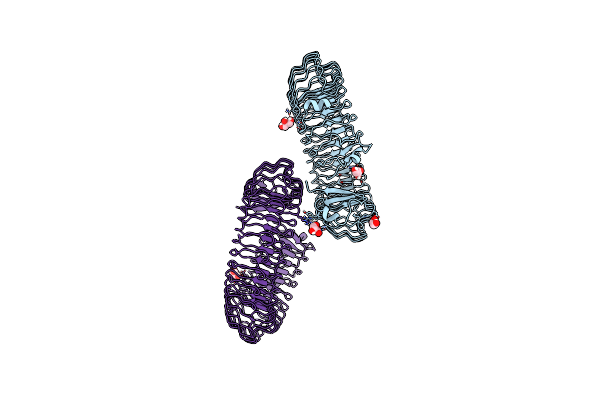
Organism: Catenovulum maritimum
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2021-03-31 Classification: LYASE Ligands: CA |
 |
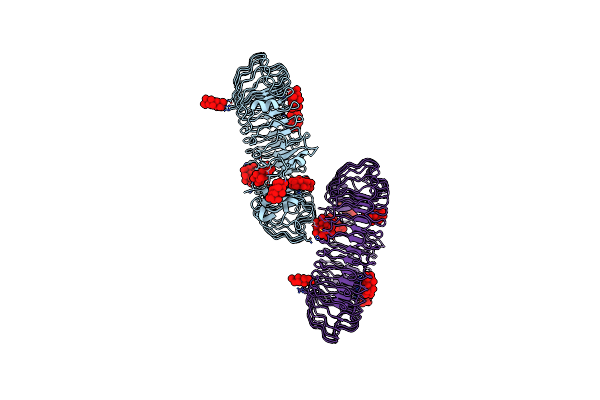
Organism: Cafeteriavirus
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2020-08-12 Classification: UNKNOWN FUNCTION Ligands: GOL |
 |
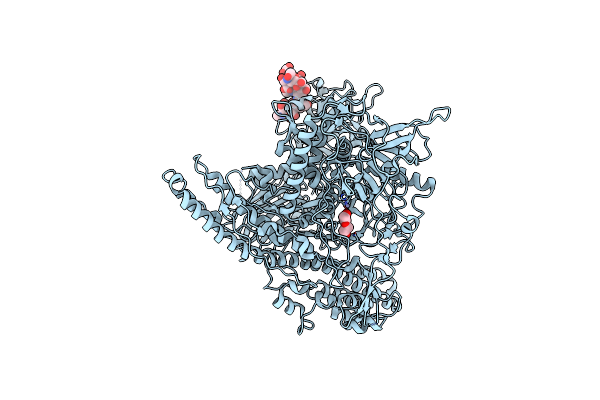
Organism: Cafeteria roenbergensis virus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2020-08-12 Classification: UNKNOWN FUNCTION Ligands: TEW |
 |
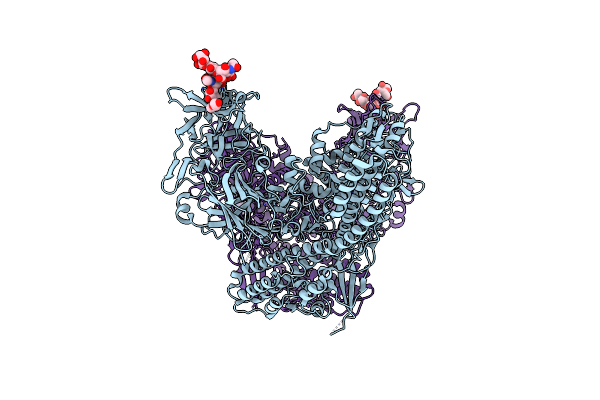
Organism: Clostridium tetani
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-06-21 Classification: TOXIN Ligands: ZN, PEG |
 |
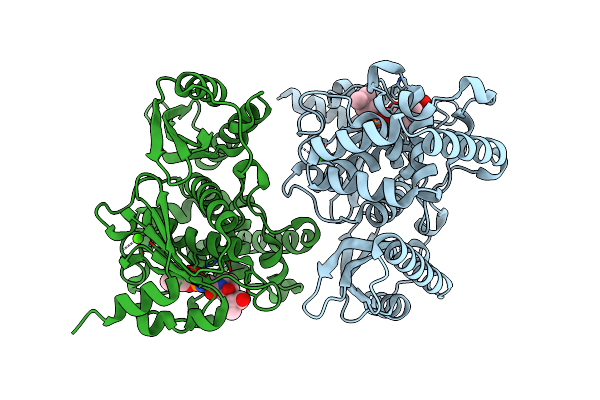
Organism: Clostridium tetani
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-06-21 Classification: TOXIN Ligands: ZN, PEG |
 |
Crystal Structure Of The Peptidase Domain Of Collagenase T From Clostridium Tetani Complexed With The Peptidic Inhibitor Isoamyl- Phosphonyl-Gly-Pro-Ala At 2.05 Angstrom Resolution.
Organism: Clostridium tetani, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2013-06-05 Classification: HYDROLASE/INHIBITOR Ligands: ZN, CA |
 |
Crystal Structure Of The Peptidase Domain Of Collagenase T From Clostridium Tetani At 1.69 Angstrom Resolution.
Organism: Clostridium tetani
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2013-06-05 Classification: HYDROLASE Ligands: ZN, CA |
 |
Crystal Structure Of The N-Terminal Domain Of The Minor Coat Protein Piii From Ctxphi
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2012-08-29 Classification: PROTEIN BINDING |