Search Count: 394
 |
Cryoem Structure Of De Novo Antibody Fragment Scfv 6 With C. Difficile Toxin B (Tcdb)
Organism: Clostridioides difficile, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: DE NOVO PROTEIN |
 |
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: K |
 |
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TRANSCRIPTION |
 |
X-Ray Structure Of Clostridioides Difficile Autolysin Acd33800 Catalytic Domain
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: ANTIMICROBIAL PROTEIN |
 |
De Novo Designed Minibinder Complexed With Clostridioides Difficile Toxin B
Organism: Clostridioides difficile, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: TOXIN |
 |
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: TOXIN Ligands: CA |
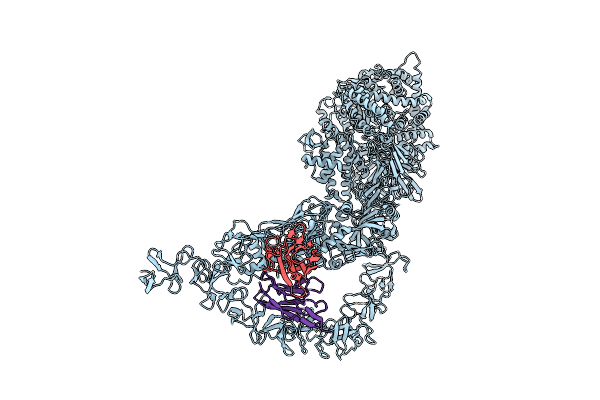 |
Organism: Clostridioides difficile, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: TRANSFERASE,TOXIN/IMMUNE SYSTEM |
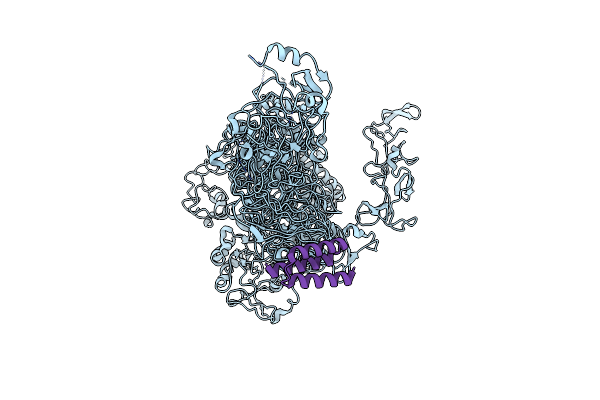 |
Organism: Synthetic construct, Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: TOXIN |
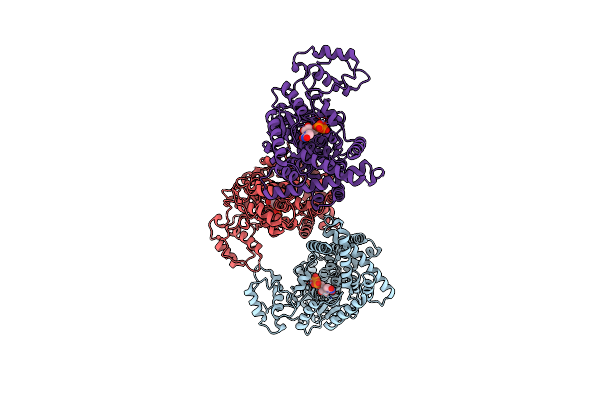 |
The Crystal Structure Of Glucosyltransferase Tcdb From Clostridioides Difficile
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:3.95 Å Release Date: 2025-01-29 Classification: TRANSFERASE Ligands: MN, UDP |
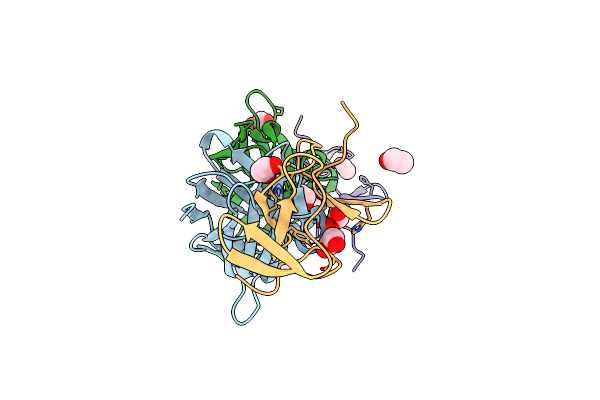 |
Crystal Structure Of The C. Difficile Toxin A Crops Domain Fragment 2639-2707 Bound To C4.2 Nanobody
Organism: Camelus dromedarius, Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-12-18 Classification: TOXIN/IMMUNE SYSTEM Ligands: EDO |
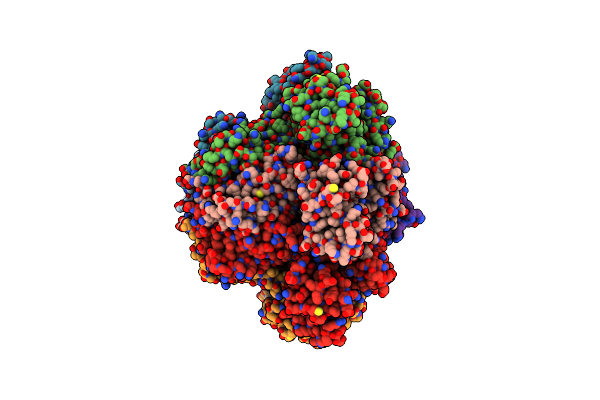 |
Structure Of Clostridioides Difficile Component A (50-463) In Complex With A Cdtb Oligomer
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-11-27 Classification: TRANSFERASE Ligands: CA |
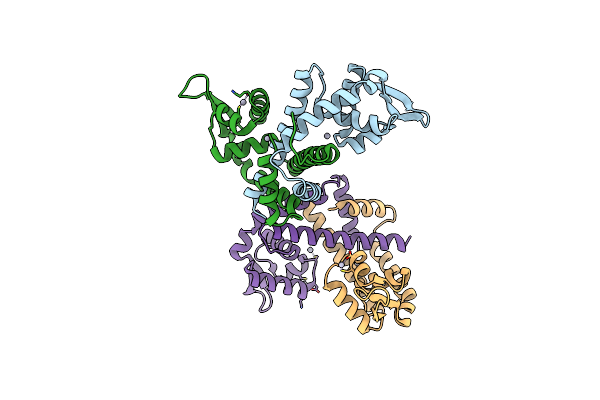 |
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-11-20 Classification: TRANSCRIPTION Ligands: HG |
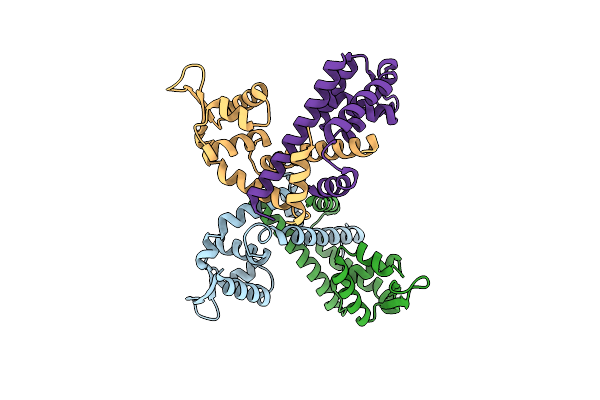 |
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-11-20 Classification: TRANSCRIPTION |
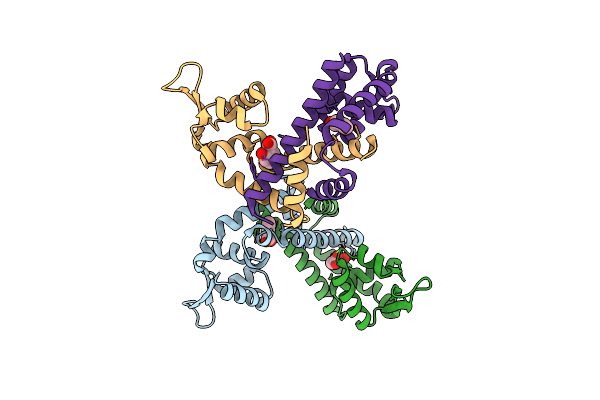 |
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2024-11-20 Classification: TRANSCRIPTION Ligands: SAL |
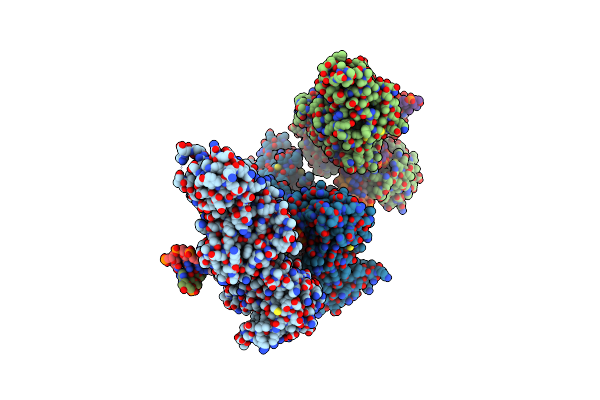 |
Cama Adenine Methyltransferase Complexed To Cognate Substrate Dna And Containing Quinoline-Based Sgi-1027 Analog 455
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2024-09-25 Classification: TRANSFERASE, DNA BINDING PROTEIN/DNA Ligands: A1AC7 |
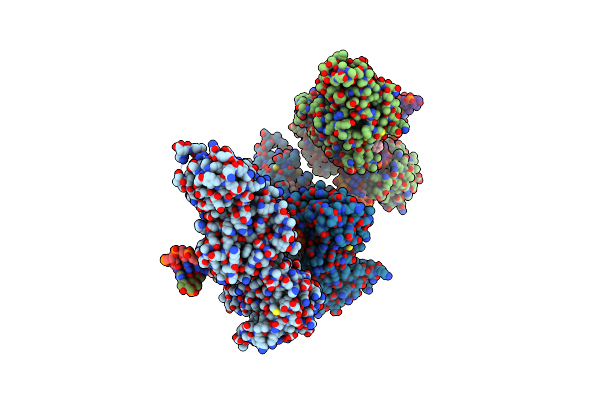 |
Cama Adenine Methyltransferase Complexed To Cognate Substrate Dna And Containing Quinoline-Based Sgi-1027 Analog 455 And Inhibitor Mc4741
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:3.18 Å Release Date: 2024-09-25 Classification: Transferase, DNA Binding Protein/DNA Ligands: Q8Y, A1AC7 |
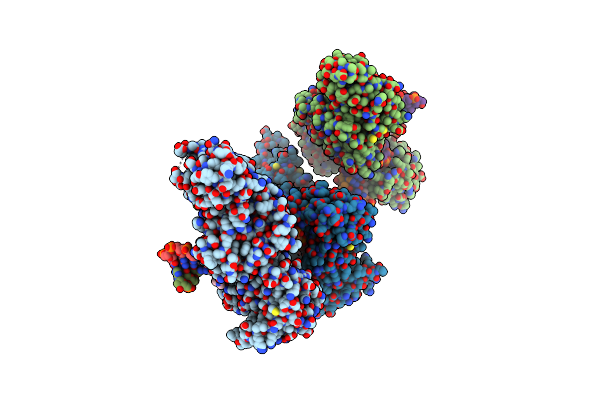 |
Cama Adenine Methyltransferase Complexed To Cognate Substrate Dna And Containing Quinoline-Based Sgi-1027 Analog 462
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2024-09-25 Classification: TRANSFERASE, DNA BINDING PROTEIN/DNA Ligands: A1ADB, 0PY |
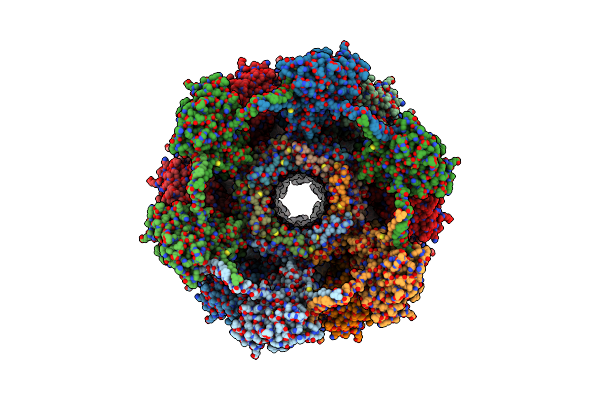 |
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
 |
Cryoem Structure Of Diffocin - Precontracted - Baseplate - Focused Refinement On Triplex Region
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Clostridioides difficile
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRUS LIKE PARTICLE |

