Search Count: 160
 |
Crystal Structure Of Cryptosporidium Parvum - Trypanosoma Cruzi Mutant Lysyl Trna Synthetase In Complex With Inhibitor
Organism: Cryptosporidium parvum iowa
Method: X-RAY DIFFRACTION Release Date: 2025-03-05 Classification: LIGASE Ligands: LYS, A1H4X, GOL, TRS, SO4 |
 |
Organism: Cryptosporidium parvum iowa
Method: X-RAY DIFFRACTION Release Date: 2024-11-06 Classification: LIGASE Ligands: LYS, A1H4W, TRS, GOL |
 |
Organism: African swine fever virus pig/kenya/ken-50/1950
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: ISOMERASE/DNA Ligands: MG, ASW |
 |
Organism: African swine fever virus pig/kenya/ken-50/1950
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: ISOMERASE |
 |
Organism: African swine fever virus pig/kenya/ken-50/1950
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: ISOMERASE |
 |
Organism: African swine fever virus pig/kenya/ken-50/1950
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2024-09-18 Classification: ISOMERASE Ligands: ATP |
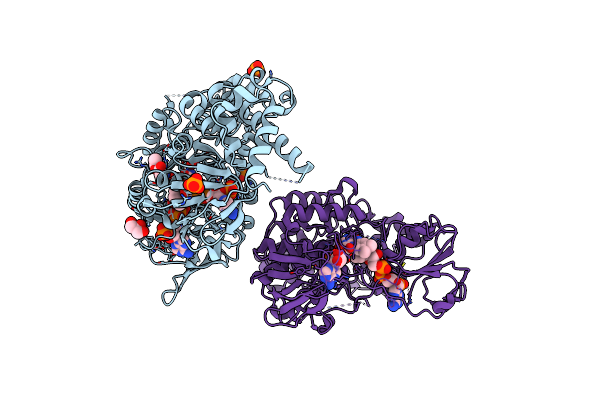 |
Crystal Structure Of Lbma Ox-Acp Didomain In Complex With Nadp And Ethyl Glycinate From The Lobatamide Pks (Gynuella Sunshinyii)
Organism: Gynuella sunshinyii
Method: X-RAY DIFFRACTION Resolution:3.07 Å Release Date: 2023-05-31 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD, NAP, OXY, GEE, GOL, PO4 |
 |
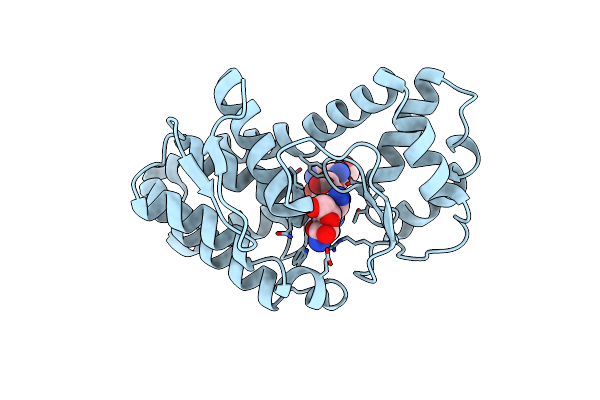
Complex Of Gynuella Sunshinyii Gh46 Chitosanase Gscsn46A With Chitotetraose
Organism: Gynuella sunshinyii yc6258
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2021-05-26 Classification: SUGAR BINDING PROTEIN Ligands: GCS |
 |
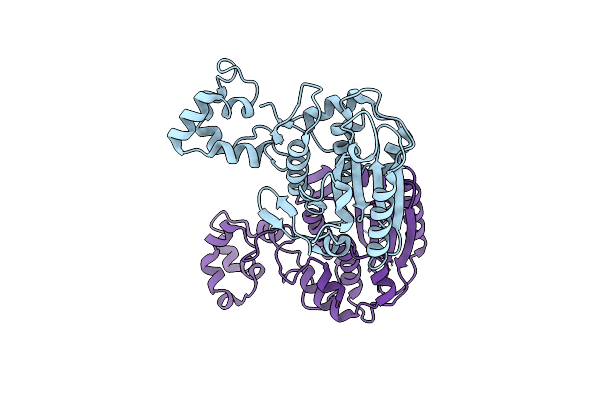
Complex Of Gynuella Sunshinyii Gh46 Chitosanase Gscsn46A E19A With Chitopentaose
Organism: Gynuella sunshinyii yc6258
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: GCS |
 |
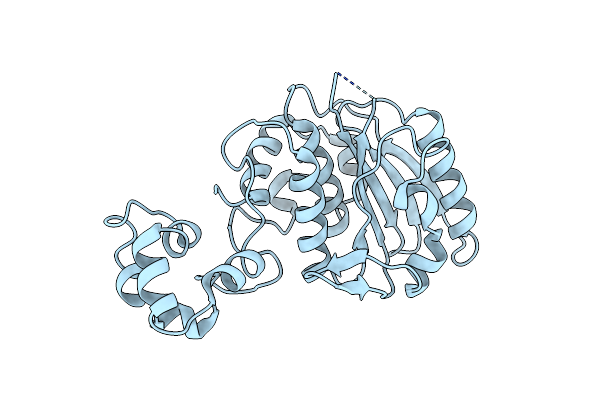
Organism: African swine fever virus pig/kenya/ken-50/1950
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2020-02-26 Classification: HYDROLASE |
 |
Organism: African swine fever virus pig/kenya/ken-50/1950
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2020-02-26 Classification: HYDROLASE |
 |
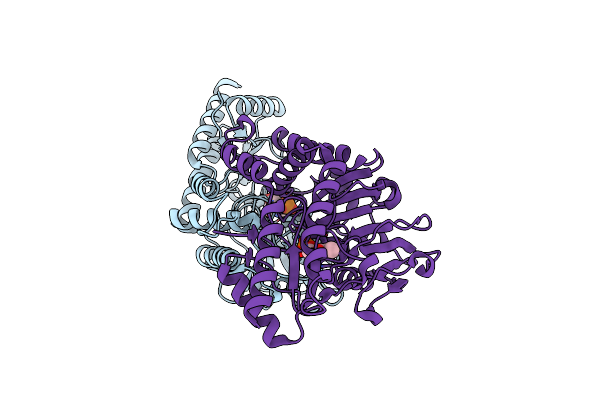
Structural Basis Of L-Phosphoserine Binding To Bacillus Alcalophilus Phosphoserine Aminotransferase
Organism: Bacillus alcalophilus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2013-05-01 Classification: TRANSFERASE Ligands: SEP, PLP, CL, NA |
 |
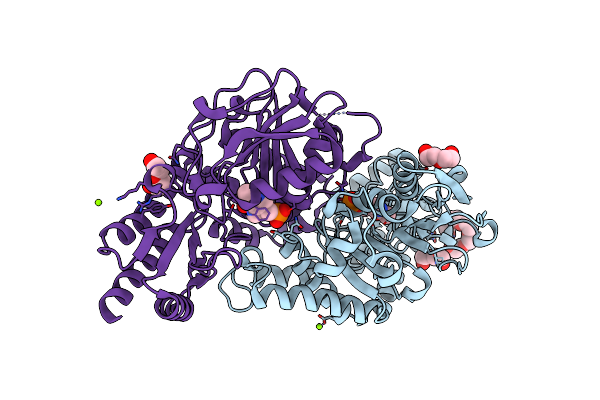
Structural Basis Of L-Phosphoserine Binding To Bacillus Alcalophilus Phosphoserine Aminotransferase
Organism: Bacillus alcalophilus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-05-01 Classification: TRANSFERASE Ligands: PLP, CL |
 |
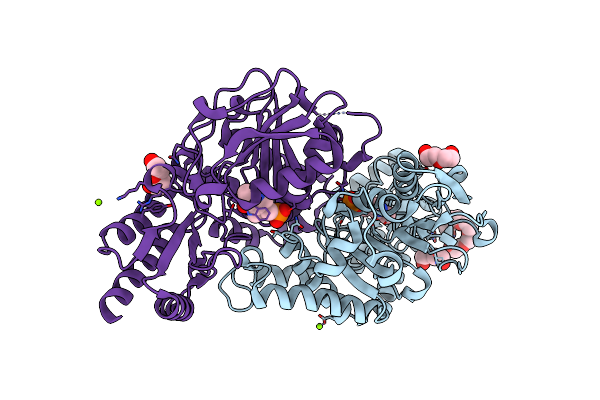
Radiation Damage Of The Schiff Base In Phosphoserine Aminotransferase (Structure A)
Organism: Bacillus alcalophilus
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2005-05-19 Classification: TRANSFERASE Ligands: PLP, MG, CL, 1PE, PEG |
 |
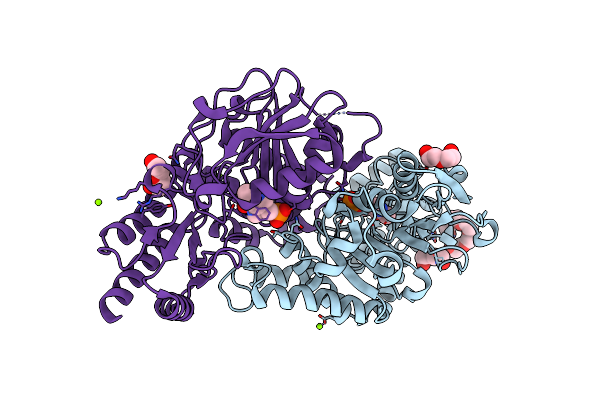
Radiation Damage Of The Schiff Base In Phosphoserine Aminotransferase (Structure B)
Organism: Bacillus alcalophilus
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2005-05-19 Classification: TRANSFERASE Ligands: PLP, MG, CL, 1PE, PEG |
 |
Radiation Damage Of The Schiff Base In Phosphoserine Aminotransferase (Structure C)
Organism: Bacillus alcalophilus
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2005-05-19 Classification: TRANSFERASE Ligands: PLP, MG, CL, 1PE, PEG |
 |
Radiation Damage Of The Schiff Base In Phosphoserine Aminotransferase (Structure D)
Organism: Bacillus alcalophilus
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2005-05-19 Classification: TRANSFERASE Ligands: PLP, MG, CL, 1PE, PEG |
 |
Radiation Damage Of The Schiff Base In Phosphoserine Aminotransferase (Structure E)
Organism: Bacillus alcalophilus
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2005-05-19 Classification: TRANSFERASE Ligands: PLP, MG, CL, 1PE, PEG |
 |
Radiation Damage Of The Schiff Base In Phosphoserine Aminotransferase (Structure F)
Organism: Bacillus alcalophilus
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2005-05-19 Classification: TRANSFERASE Ligands: PLP, MG, CL, 1PE, PEG |
 |
Radiation Damage Of The Schiff Base In Phosphoserine Aminotransferase (Structure G)
Organism: Bacillus alcalophilus
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2005-05-19 Classification: TRANSFERASE Ligands: PLP, MG, CL, 1PE, PEG |

