Search Count: 290
 |
Cereblon Isoform 4 In Complex With Novel Benzamide-Type Cereblon Binder 11C
Organism: Magnetospirillum gryphiswaldense msr-1 v2
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2023-11-15 Classification: SIGNALING PROTEIN Ligands: ZN, W26, PO4 |
 |
Mosquitocidal Cry11Aa Determined At Ph 7 From Naturally-Occurring Nanocrystals By Serial Femtosecond Crystallography
Organism: Bacillus thuringiensis serovar israelensis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-07-27 Classification: TOXIN |
 |
Mosquitocidal Cry11Aa-Y449F Determined At Ph 7 From Naturally-Occurring Nanocrystals By Serial Femtosecond Crystallography
Organism: Bacillus thuringiensis serovar israelensis
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2022-07-27 Classification: TOXIN |
 |
Mosquitocidal Cry11Aa-E583Q Determined At Ph 7 From Naturally-Occurring Nanocrystals By Serial Femtosecond Crystallography
Organism: Bacillus thuringiensis serovar israelensis
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2022-07-27 Classification: TOXIN |
 |
Mosquitocidal Cry11Aa-F17Y Determined At Ph 7 From Naturally-Occurring Nanocrystals By Serial Femtosecond Crystallography
Organism: Bacillus thuringiensis serovar israelensis
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2022-07-27 Classification: TOXIN |
 |
Native Structure Of Mosquitocidal Cyt1A Protoxin Obtained By Serial Femtosecond Crystallography On In Vivo Grown Crystals At Ph 7
Organism: Bacillus thuringiensis subsp. israelensis
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2020-10-14 Classification: TOXIN |
 |
Structure Of Mosquitocidal Cyt1A Protoxin Obtained By Serial Femtosecond Crystallography On In Vivo Grown Crystals Soaked With Dtt At Ph 7
Organism: Bacillus thuringiensis subsp. israelensis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-10-14 Classification: TOXIN |
 |
Structure Of Mosquitocidal Cyt1Aa Protoxin Obtained By Serial Femtosecond Crystallography On In Vivo Grown Crystals At Ph 10
Organism: Bacillus thuringiensis subsp. israelensis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-10-14 Classification: TOXIN Ligands: CA |
 |
Structure Of The C7S Mutant Of Mosquitocidal Cyt1A Protoxin Obtained By Serial Femtosecond Crystallography On In Vivo Grown Crystals At Ph 7
Organism: Bacillus thuringiensis subsp. israelensis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-10-14 Classification: TOXIN |
 |
Organism: Magnetospirillum gryphiswaldense
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2015-07-15 Classification: METAL BINDING PROTEIN Ligands: FLC, SO4 |
 |
Organism: Magnetospirillum gryphiswaldense
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-07-15 Classification: METAL BINDING PROTEIN Ligands: MN, EDO |
 |
Organism: Magnetospirillum gryphiswaldense
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2015-07-15 Classification: METAL BINDING PROTEIN Ligands: FLC, SO4 |
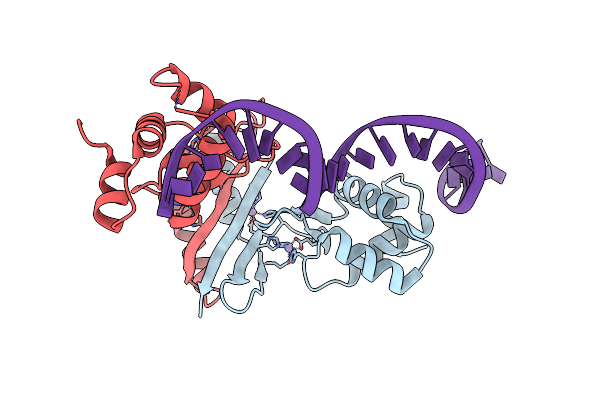 |
Crystal Structure Of Magnetospirillum Gryphiswaldense Msr-1 Fur-Mn2+-E. Coli Fur Box
Organism: Magnetospirillum gryphiswaldense
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2015-07-15 Classification: METAL BINDING PROTEIN/DNA Ligands: MN |
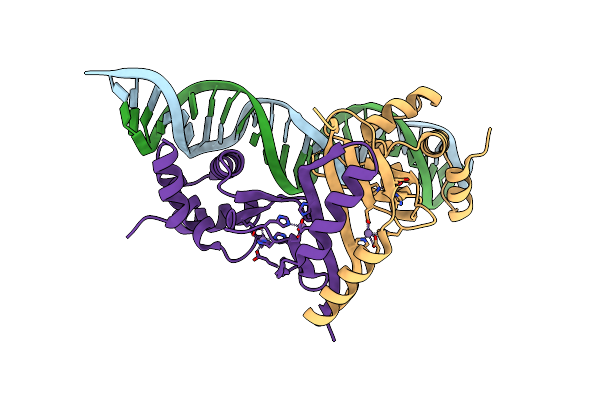 |
Crystal Structure Of Magnetospirillum Gryphiswaldense Msr-1 Semet-Fur-Mn2+-Feoab1 Operator
Organism: Magnetospirillum gryphiswaldense
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2015-07-15 Classification: METAL BINDING PROTEIN/DNA Ligands: MN |
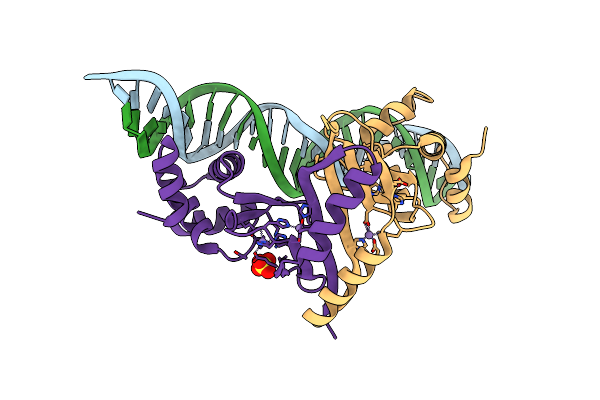 |
Crystal Structure Of Magnetospirillum Gryphiswaldense Msr-1 Fur-Mn2+-Feoab1 Operator
Organism: Magnetospirillum gryphiswaldense
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-07-15 Classification: METAL BINDING PROTEIN/DNA Ligands: MN, SO4 |
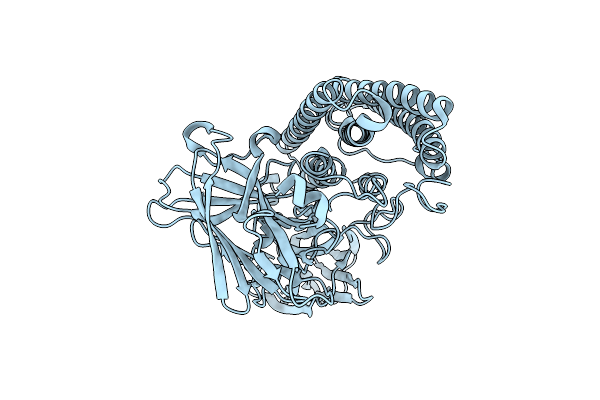 |
Organism: Bacillus thuringiensis serovar israelensis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-12-10 Classification: TOXIN |
 |
Organism: Bacillus thuringiensis
Method: ELECTRON MICROSCOPY Release Date: 2014-02-19 Classification: STRUCTURAL PROTEIN Ligands: GDP |
 |
Organism: Bacillus thuringiensis
Method: ELECTRON MICROSCOPY Release Date: 2014-02-19 Classification: STRUCTURAL PROTEIN |
 |
Organism: Gallus gallus, Micrurus tener tener
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2014-02-19 Classification: TRANSPORT PROTEIN/TOXIN Ligands: NAG, CL, NA, P6G |
 |
Structure Of Acid-Sensing Ion Channel In Complex With Snake Toxin And Amiloride
Organism: Gallus gallus, Micrurus tener tener
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2014-02-19 Classification: TRANSPORT PROTEIN/TOXIN Ligands: NAG, CL, NA, P6G, AMR |

