Search Count: 97
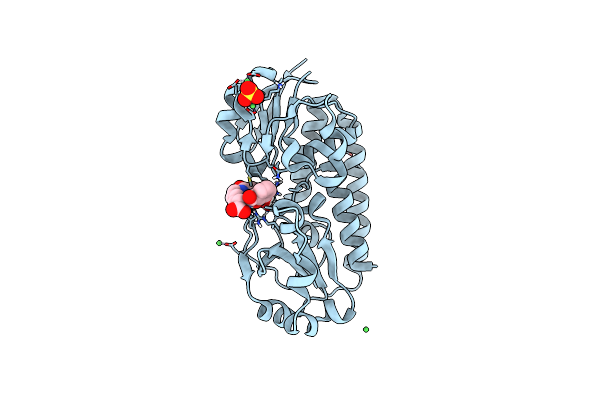 |
X-Ray Structure Of The Ceue Homologue From Parageobacillus Thermoglucosidasius - Azotochelin Complex
Organism: Parageobacillus thermoglucosidasius
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2023-07-12 Classification: METAL TRANSPORT Ligands: FE, 95B, NI, SO4 |
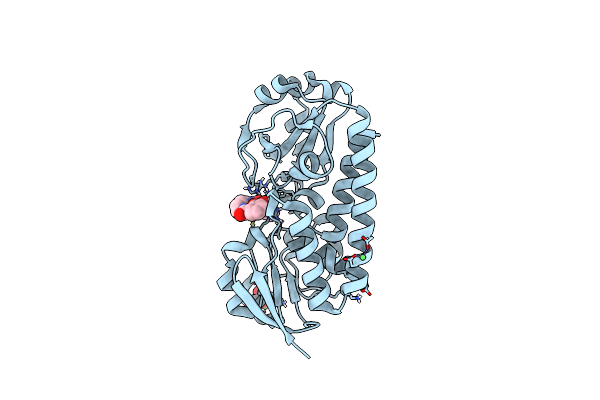 |
X-Ray Structure Of The Ceue Homologue From Parageobacillus Thermoglucosidasius - 5Licam Complex.
Organism: Parageobacillus thermoglucosidasius
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2023-07-12 Classification: METAL TRANSPORT Ligands: FE, 5LC, NI, SO4 |
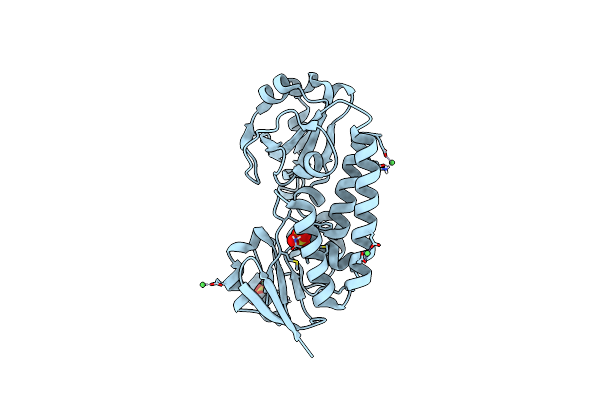 |
X-Ray Structure Of The Ceue Homologue From Parageobacillus Thermoglucosidasius - Apo Form
Organism: Parageobacillus thermoglucosidasius
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2023-07-12 Classification: METAL TRANSPORT Ligands: NI, SO4 |
 |
Crystal Structure Of L-2,4-Diketo-3-Deoxyrhamnonate Hydrolase From Sphingomonas Sp. (Apo-Form)
Organism: Sphingomonas sp. ska58
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2023-02-08 Classification: HYDROLASE Ligands: MG |
 |
Crystal Structure Of L-2,4-Diketo-3-Deoxyrhamnonate Hydrolase From Sphingomonas Sp. (Pyruvate Bound-Form)
Organism: Sphingomonas sp. ska58
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2023-02-08 Classification: HYDROLASE Ligands: MG, PYR |
 |
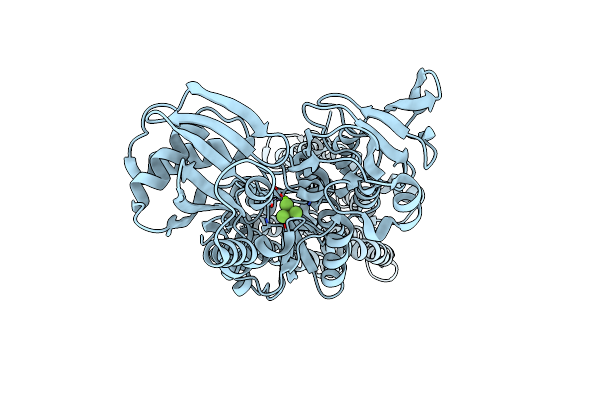
Organism: Sulfitobacter sp. (strain nas-14.1)
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2022-03-23 Classification: MEMBRANE PROTEIN Ligands: ALF, MG |
 |
Organism: Sulfitobacter sp. (strain nas-14.1)
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2022-03-23 Classification: MEMBRANE PROTEIN Ligands: BEF, MG |
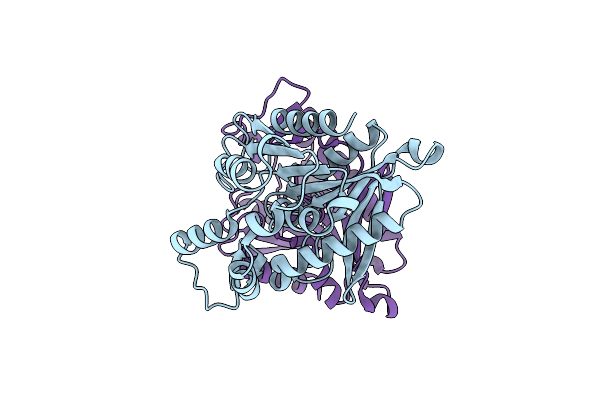 |
Crystal Structure Of The Geobacillus Thermoglucosidasius Feruloyl Esterase Gthfae
Organism: Parageobacillus thermoglucosidasius
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2022-03-09 Classification: HYDROLASE |
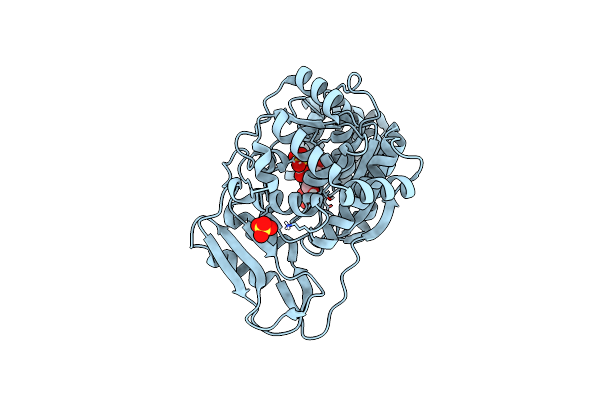 |
Crystal Structure Of Uridine Bound To Geobacillus Thermoglucosidasius Pyrimidine Nucleoside Phosphorylase Pynp
Organism: Parageobacillus thermoglucosidasius
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2021-08-25 Classification: TRANSFERASE Ligands: SO4, URI |
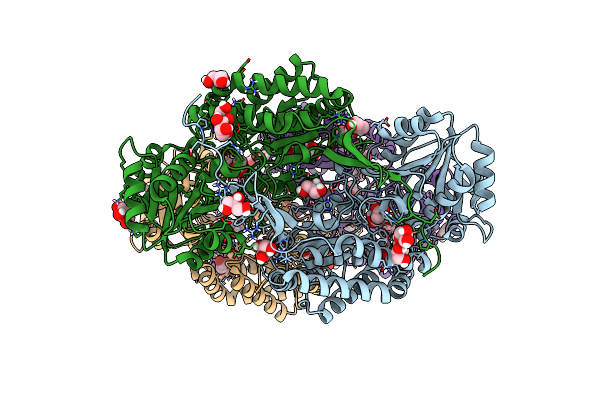 |
Crystal Structure Of An Acetylating Aldehyde Dehydrogenase From Geobacillus Thermoglucosidasius
Organism: Geobacillus thermoglucosidasius
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-09-07 Classification: OXIDOREDUCTASE Ligands: ACT, GOL, MG |
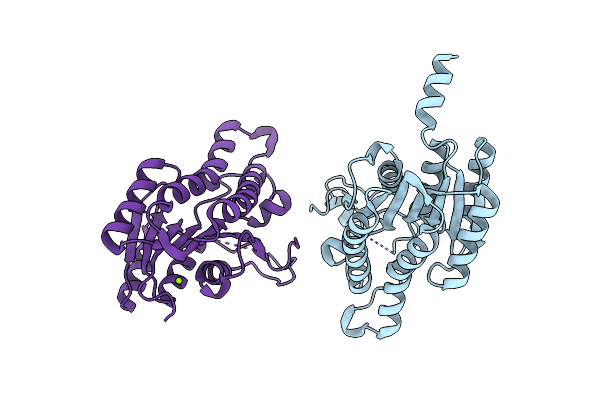 |
Crystal Structure Of A Putative Dehydrogenase From Sulfitobacter Sp. (Cog1028) (Target Efi-513936) In Its Apo Form
Organism: Sulfitobacter sp. nas-14.1
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2015-03-25 Classification: OXIDOREDUCTASE Ligands: MG |
 |
Organism: Geobacillus thermoglucosidasius
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-05-14 Classification: HYDROLASE Ligands: SO4, NA, EDO, GOL, PEG, PGE |
 |
Organism: Geobacillus thermoglucosidasius
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2014-05-14 Classification: HYDROLASE |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Sulfitobacter Sp. Nas-14.1 (Target Efi-510299) With Bound Beta-D-Galacturonate
Organism: Sulfitobacter sp. nas-14.1
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-05-14 Classification: TRANSPORT PROTEIN Ligands: CL, GTR |
 |
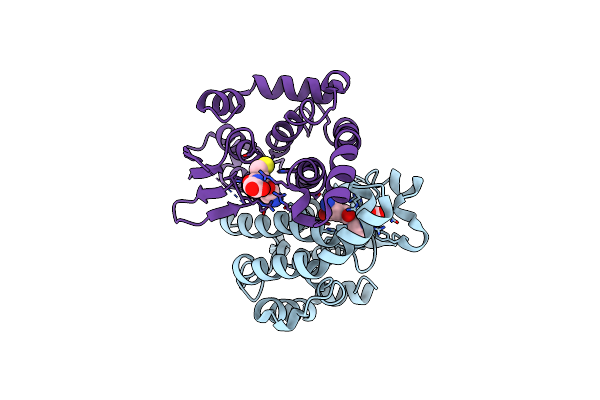
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Sulfitobacter Sp. Nas-14.1, Target Efi-510292, With Bound Alpha-D-Taluronate
Organism: Sulfitobacter sp. nas-14.1
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-01-22 Classification: TRANSPORT PROTEIN Ligands: X1X |
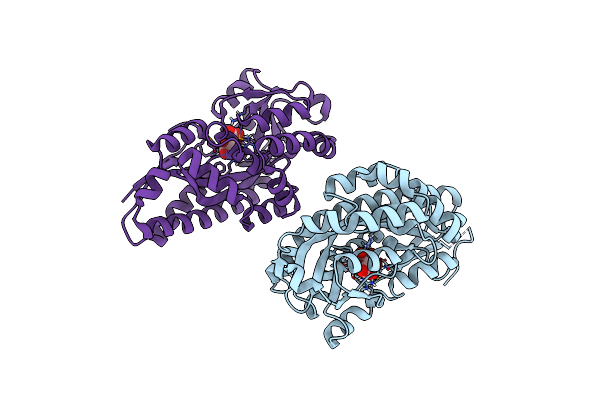 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Sulfitobacter Sp. Nas-14.1, Target Efi-510292, With Bound Alpha-D-Manuronate
Organism: Sulfitobacter sp.
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-01-08 Classification: SOLUTE-BINDING PROTEIN Ligands: MAV |
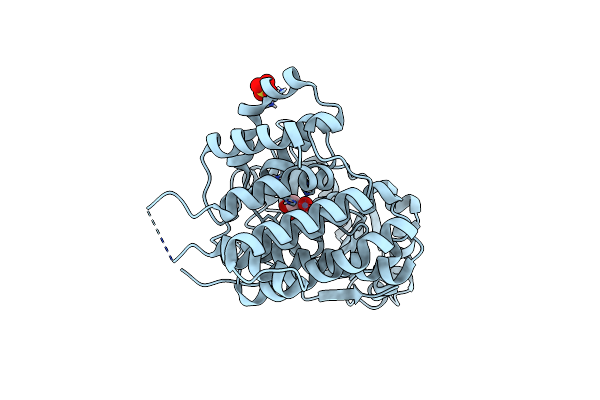 |
Structure Of The Alcohol Dehydrogenase (Adh) Domain Of A Bifunctional Adhe Dehydrogenase From Geobacillus Thermoglucosidasius Ncimb 11955
Organism: Geobacillus thermoglucosidasius
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-10-02 Classification: OXIDOREDUCTASE Ligands: ZN, SO4, GOL |
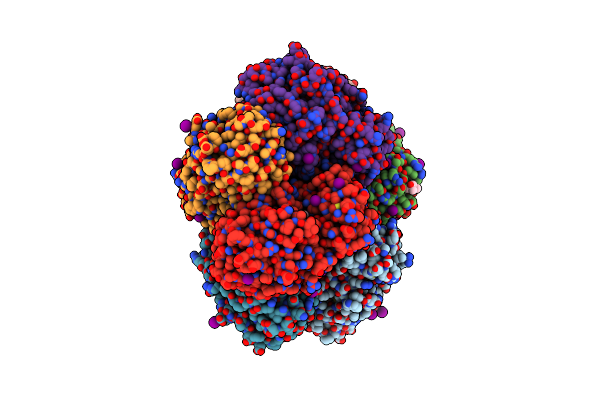 |
Crystal Structure Of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis Htcc2601, With Bound Mg And L-4-Hydroxyproline Betaine (Betonicine)
Organism: Pelagibaca bermudensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-10-10 Classification: ISOMERASE Ligands: MG, NI, 0XW, IOD, MPD |
 |
Crystal Structure Of Glutahtione Transferase Homolog From Sulfitobacter, Target Efi-501084, With Bound Glutathione
Organism: Sulfitobacter
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2012-08-29 Classification: TRANSFERASE Ligands: CL, GSH |
 |
Organism: Bacillus weihenstephanensis, Sulfitobacter sp. nas-14.1
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2012-05-02 Classification: TRANSPORT PROTEIN |

