Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
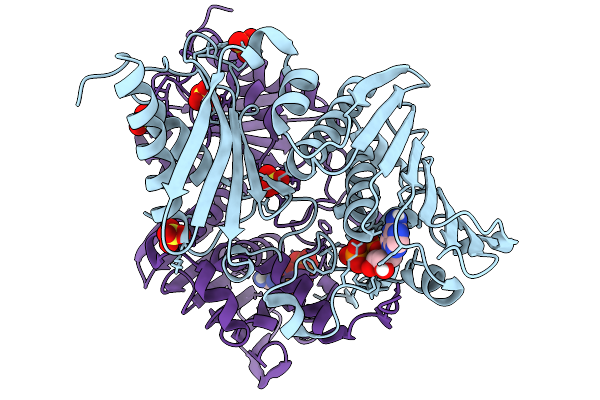 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: DNA BINDING PROTEIN Ligands: ADP, MG, SO4 |
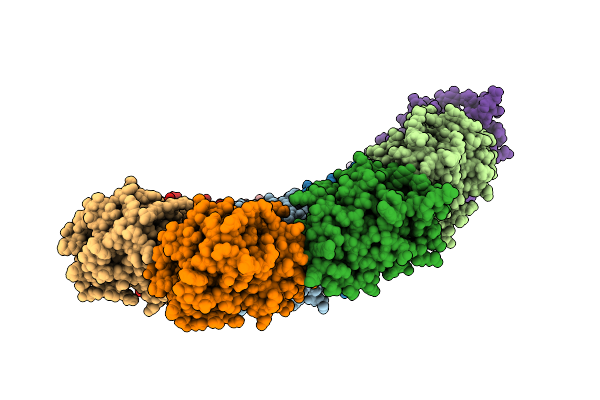 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: ANTIVIRAL PROTEIN |
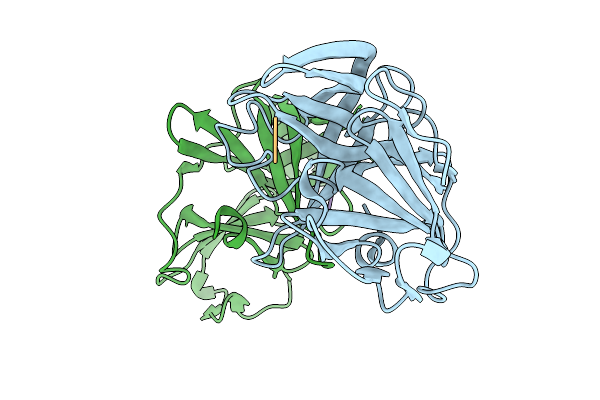 |
Organism: Norovirus, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: VIRAL PROTEIN/INHIBITOR |
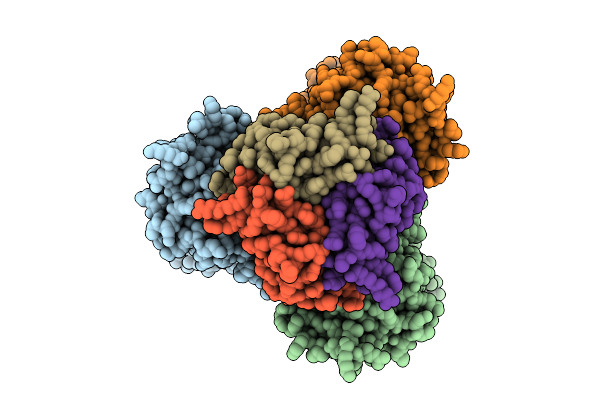 |
Cryo-Em Structure Of Bicarbonate Transporter Sbta In Complex With Pii-Like Signaling Protein Sbtb (T-Loop Truncation) From Synechocystis Sp. Pcc 6803
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: TRANSPORT PROTEIN Ligands: BCT |
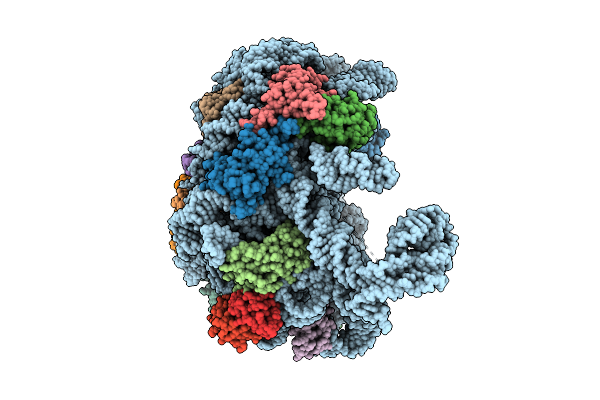 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RIBOSOME |
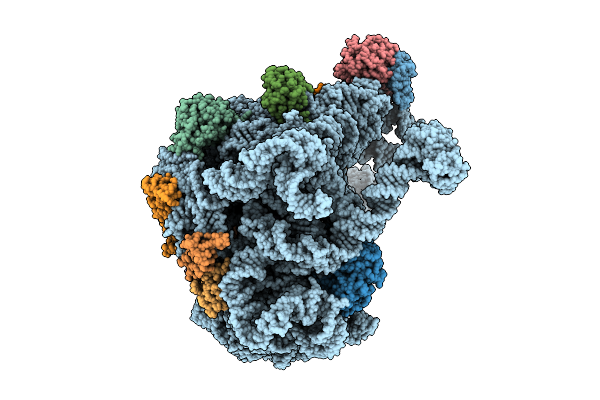 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RIBOSOME |
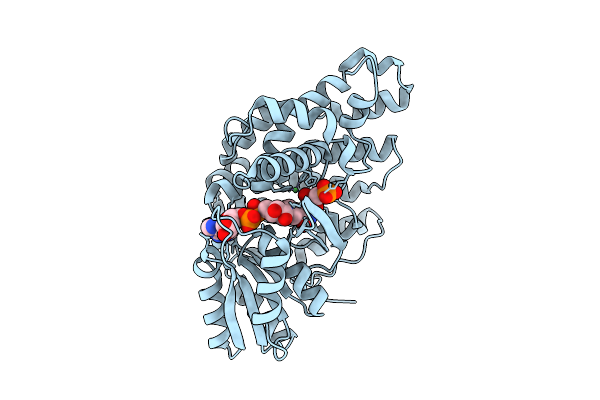 |
Crystal Structure Of Ni2+ Dependent Glycerol-1-Phosphate Dehydrogenase Aram From Bacillus Subtilis
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: BIOSYNTHETIC PROTEIN Ligands: 13P, NAI, NI |
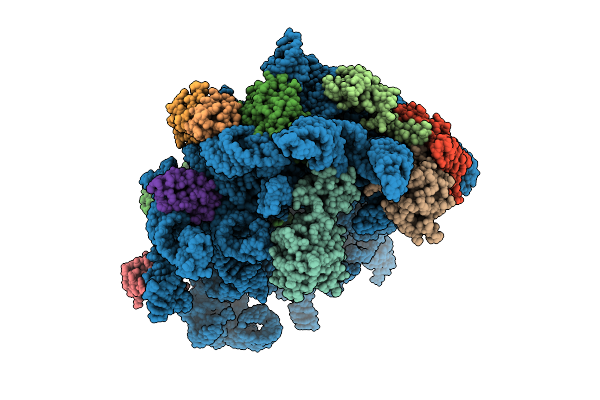 |
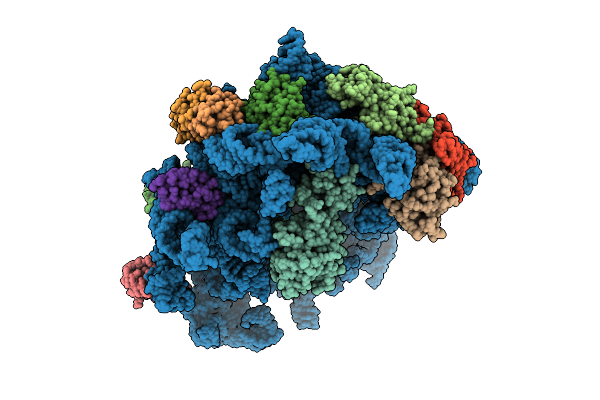
Organism: Escherichia coli k-12, Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: RIBOSOME |
 |
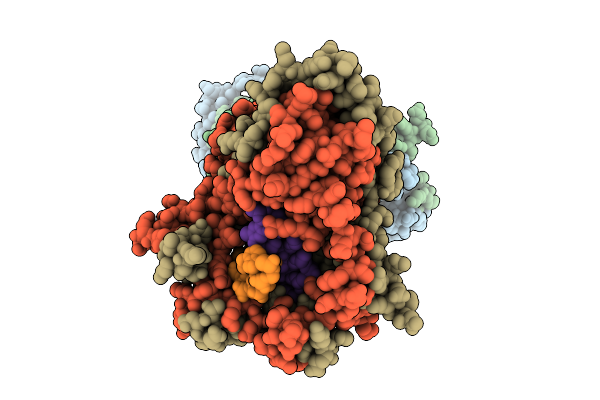
Organism: Escherichia coli k-12, Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: RIBOSOME Ligands: AN6 |
 |
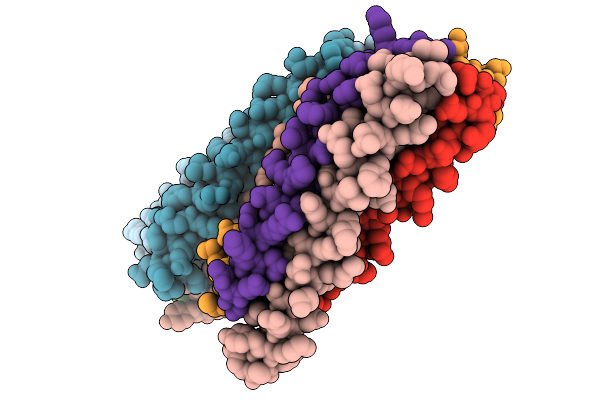
Organism: Bacillus subtilis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: DNA BINDING PROTEIN |
 |
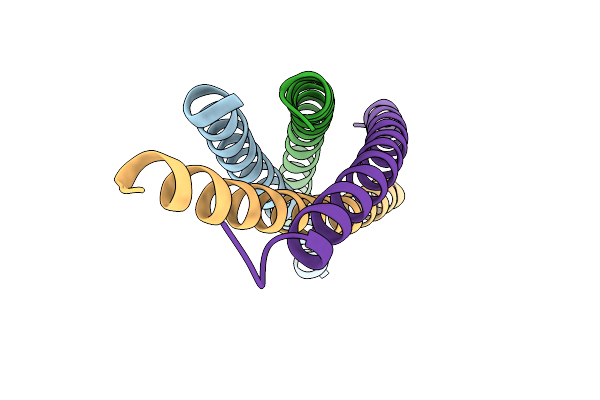
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: TRANSCRIPTION |
 |
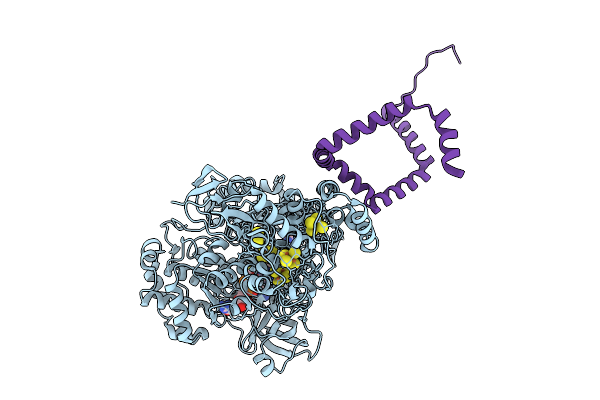
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: TRANSCRIPTION |
 |
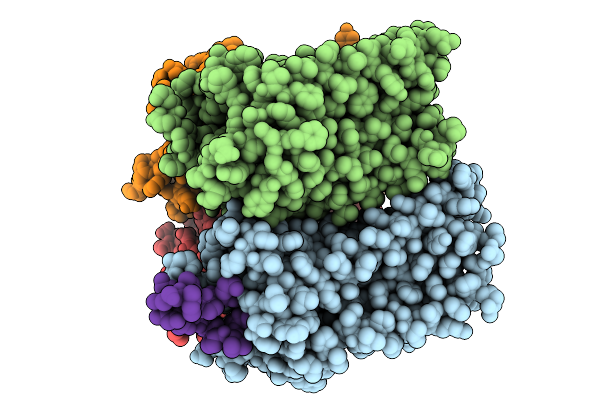
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: ELECTRON TRANSPORT Ligands: FES, SF4, MGD, 4MO |
 |
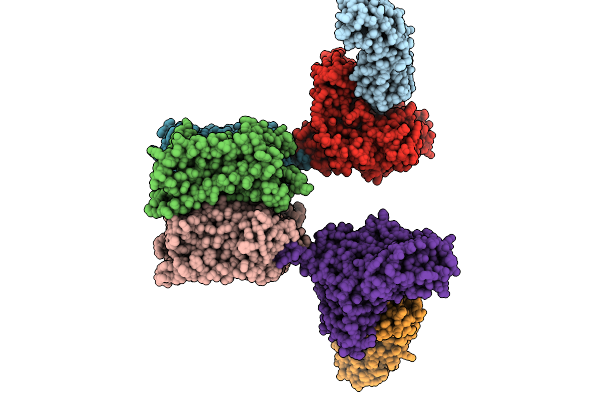
Organism: Homo sapiens, Virus-associated rnas
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens, Virus-associated rnas
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: SIGNALING PROTEIN |
 |
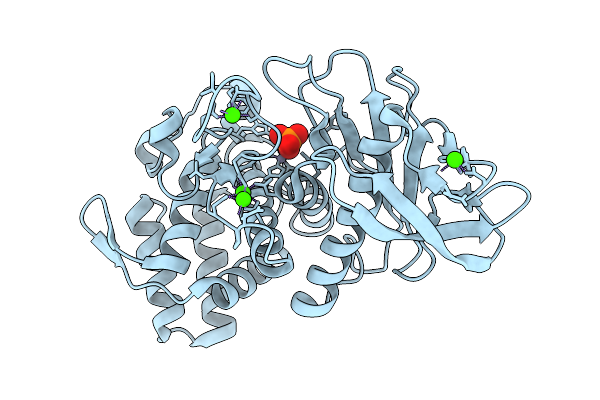
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: RNA BINDING PROTEIN |
 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: HYDROLASE Ligands: ZN, CA, PO4 |
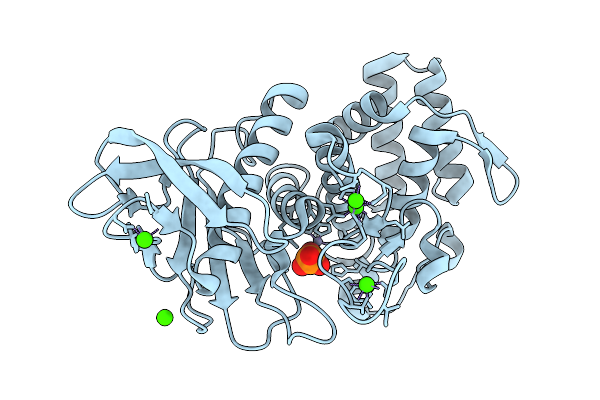 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: HYDROLASE Ligands: ZN, CA, PO4 |
 |
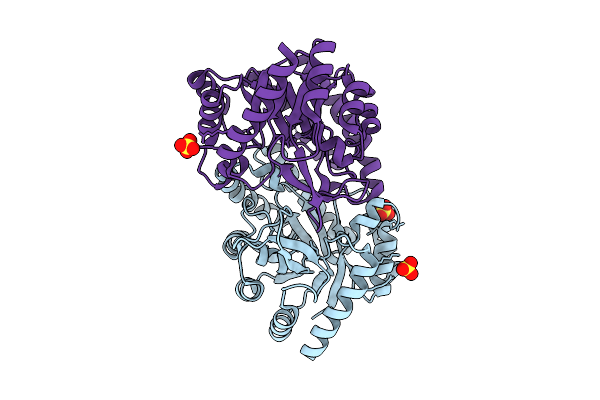
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: ISOMERASE Ligands: MN, SO4 |