Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Kroppenstedtia eburnea
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-12-25 Classification: ISOMERASE Ligands: FUD, CO, GOL |
 |
Organism: Kroppenstedtia eburnea
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2024-12-25 Classification: ISOMERASE |
 |
Organism: Kroppenstedtia eburnea
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2024-12-25 Classification: ISOMERASE |
 |
Organism: Thermus phage g20c
Method: X-RAY DIFFRACTION Resolution:2.97 Å Release Date: 2022-11-02 Classification: VIRAL PROTEIN |
 |
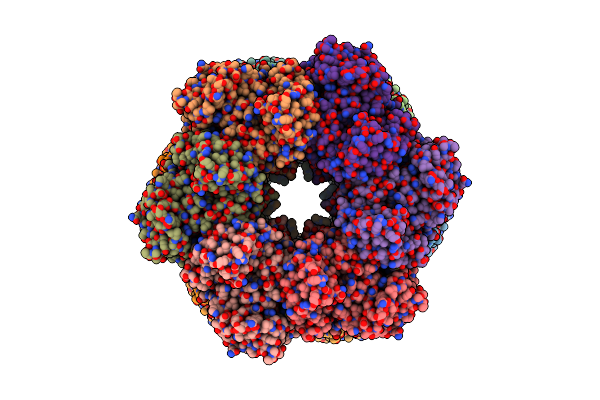
Structure Of Paenibacillus Polymyxa Gs Bound To Met-Sox-P-Adp (Transition State Complex) To 1.98 Angstom
Organism: Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2022-06-29 Classification: LIGASE Ligands: ADP, P3S, MG |
 |
Organism: Paenibacillus polymyxa
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: BIOSYNTHETIC PROTEIN, LIGASE Ligands: MG, GLN |
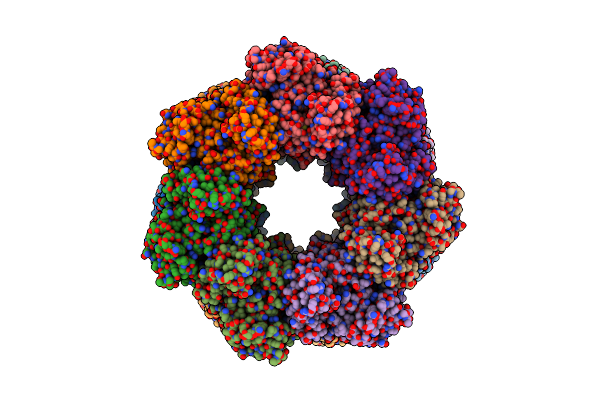 |
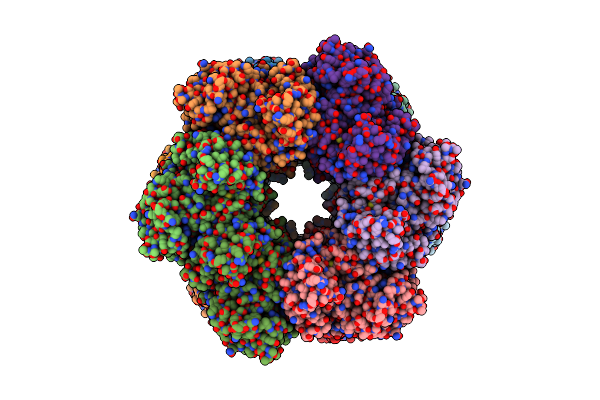
Organism: Paenibacillus polymyxa
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: BIOSYNTHETIC PROTEIN, LIGASE Ligands: MG, GLN |
 |
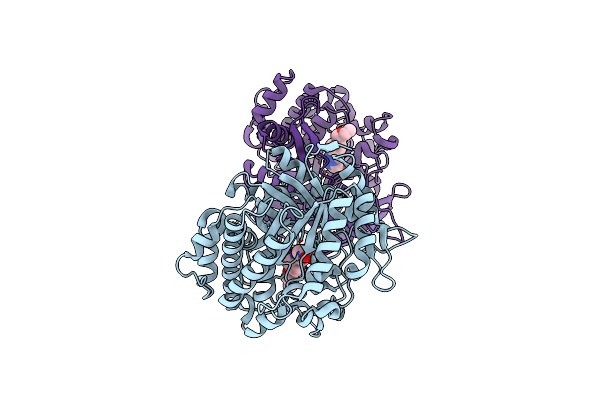
Organism: Paenibacillus polymyxa
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: BIOSYNTHETIC PROTEIN Ligands: MG |
 |
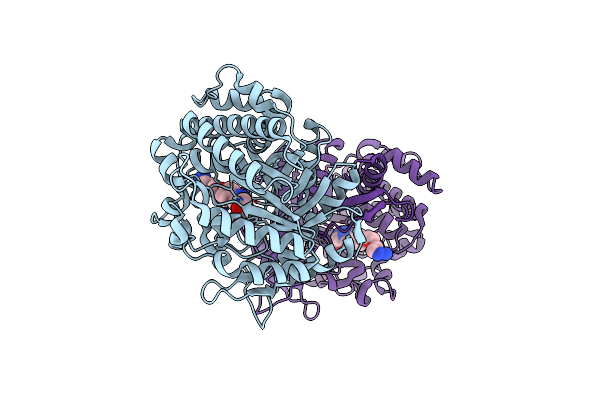
Structure Of Beta-Glucosidase A From Paenibacillus Polymyxa Complexed With Multivalent Inhibitors.
Organism: Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2019-07-03 Classification: HYDROLASE Ligands: JJW |
 |
Structure Of Beta-Glucosidase A From Paenibacillus Polymyxa Complexed With A Monovalent Inhibitor
Organism: Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2019-07-03 Classification: HYDROLASE Ligands: JSK |
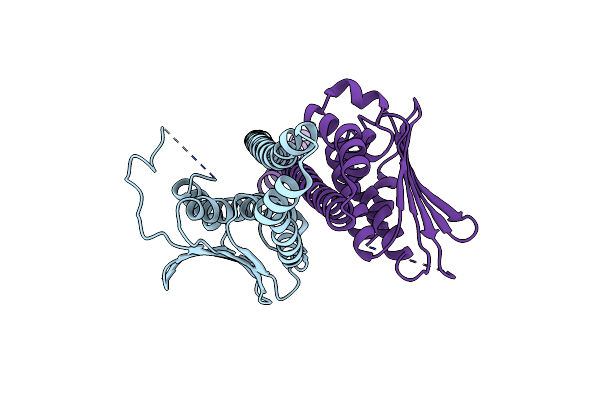 |
Chimeric Structure Of Saccharomyces Cerevisiae Gcn4 Leucine Zipper Fused To Staphylococcus Aureus Agrc Cytoplasmic Histidine Kinase Module (Dataset Anisotropically Truncated By Staraniso)
Organism: Saccharomyces cerevisiae, Staphylococcus aureus subsp. aureus z172
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2019-02-27 Classification: SIGNALING PROTEIN |
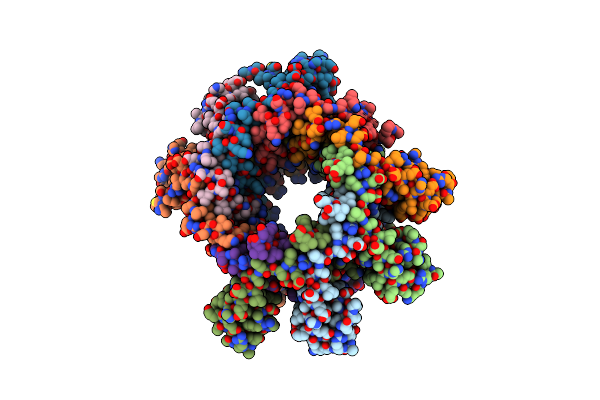 |
Crystal Structure Of A C-Terminally Truncated Small Terminase Protein From The Thermophilic Bacteriophage G20C
Organism: Thermus phage g20c
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-10-10 Classification: VIRAL PROTEIN |
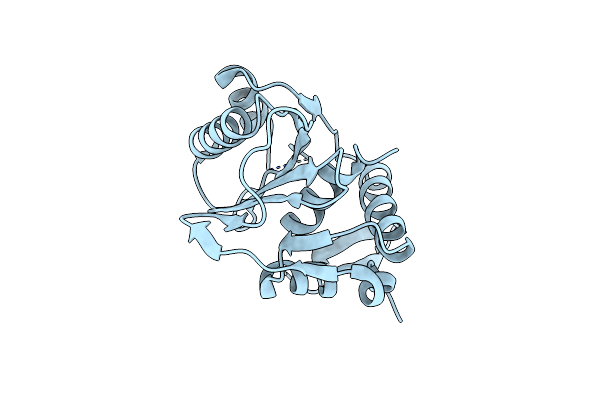 |
Crystal Structure Of The Large Terminase Nuclease From Thermophilic Phage G20C
Organism: Thermus phage g20c
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2016-10-26 Classification: VIRAL PROTEIN |
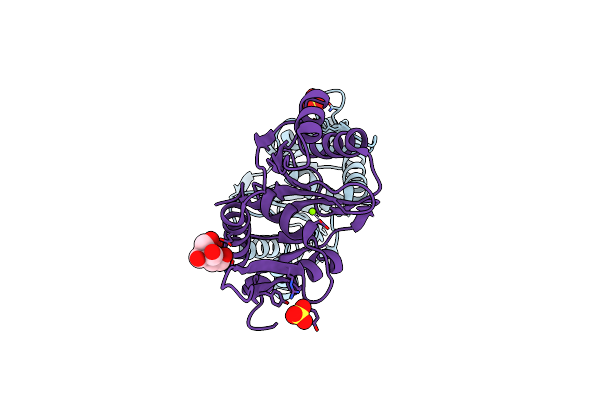 |
Crystal Structure Of The Large Terminase Nuclease From Thermophilic Phage G20C With Bound Magnesium
Organism: Thermus phage g20c
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2016-10-26 Classification: VIRAL PROTEIN Ligands: SO4, MG, BTB |
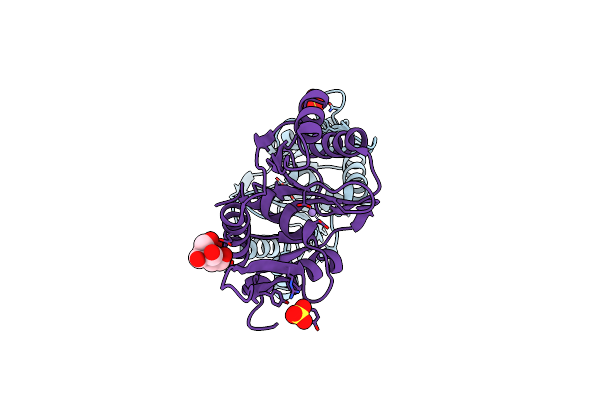 |
Crystal Structure Of The Large Terminase Nuclease From Thermophilic Phage G20C With Bound Manganese
Organism: Thermus phage g20c
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2016-10-26 Classification: VIRAL PROTEIN Ligands: SO4, MN, BTB |
 |
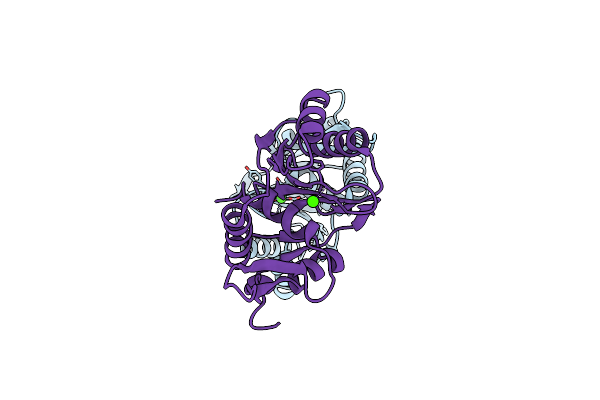
Crystal Structure Of The Large Terminase Nuclease From Thermophilic Phage G20C With Bound Cobalt
Organism: Thermus phage g20c
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-10-26 Classification: VIRAL PROTEIN Ligands: CO |
 |
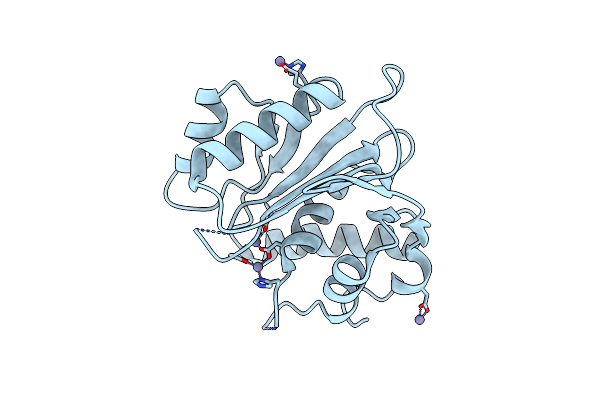
Crystal Structure Of The Large Terminase Nuclease From Thermophilic Phage G20C With Bound Calcium
Organism: Thermus phage g20c
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2016-10-26 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of The Large Terminase Nuclease From Thermophilic Phage G20C With Bound Zinc
Organism: Thermus phage g20c
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2016-10-26 Classification: VIRAL PROTEIN Ligands: ZN |
 |
Organism: Thermus phage g20c
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-02-10 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of The Catalytic Domain Of Beta-Amylase From Paenibacillus Polymyxa
Organism: Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-02-20 Classification: HYDROLASE Ligands: CA, CL, EDO, GOL, TRS, PEG |