Search Count: 34
 |
Organism: Escherichia coli str. k-12 substr. mc4100
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2015-11-25 Classification: OXIDOREDUCTASE Ligands: FCO, NI, MG, SO4, SF4, F3S, SF3, CL |
 |
Organism: Escherichia coli str. k-12 substr. mc4100
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2015-11-25 Classification: OXIDOREDUCTASE Ligands: FCO, NI, MG, SO4, SF4, F3S, SF3, LMT, CL |
 |
Organism: Escherichia coli str. k-12 substr. mc4100
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2015-11-25 Classification: OXIDOREDUCTASE Ligands: FCO, NI, MG, SO4, SF4, F3S, SF3, LMT, CL |
 |
Organism: Escherichia coli str. k-12 substr. mc4100
Method: SOLUTION NMR Release Date: 2015-05-20 Classification: ELECTRON TRANSPORT Ligands: FMN |
 |
Refined Structure Of Outer Membrane Protein X In Nanodisc By Measuring Residual Dipolar Couplings
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2015-03-18 Classification: MEMBRANE PROTEIN |
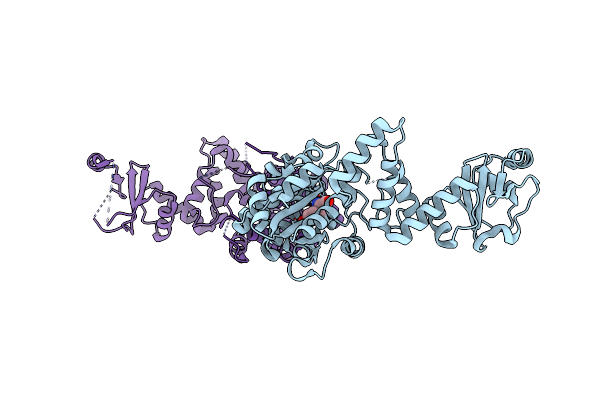 |
Engineered Tyrosyl-Trna Synthetase With The Nonstandard Amino Acid L-4,4-Biphenylalanine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2015-01-28 Classification: LIGASE Ligands: TYR |
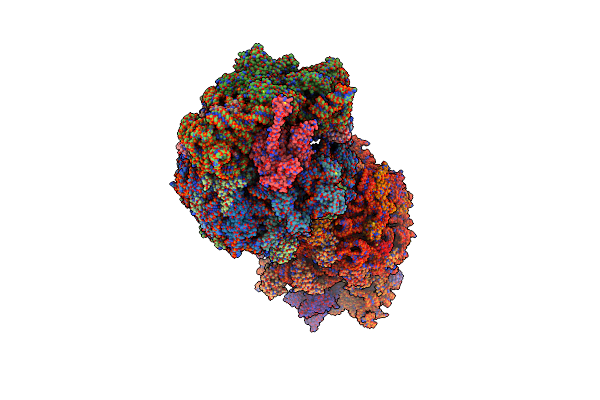 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Elongation Factor G In The Pre-Translocational State
Organism: Escherichia coli str. k-12 substr. mc4100, Thermus thermophilus, Synthetic construct, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-01-28 Classification: RIBOSOME Ligands: MG, K, ZN, SF4, GDP |
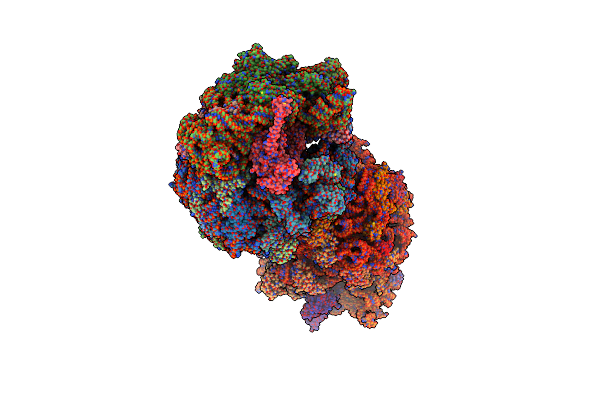 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Elongation Factor G And Fusidic Acid In The Post-Translocational State
Organism: Escherichia coli str. k-12 substr. mc4100, Thermus thermophilus, Synthetic construct, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-01-28 Classification: RIBOSOME Ligands: MG, ZN, SF4, FUA, GDP |
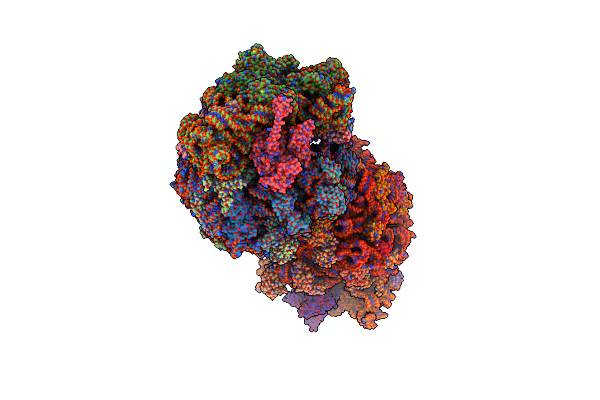 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Elongation Factor G Trapped By The Antibiotic Dityromycin
Organism: Escherichia coli str. k-12 substr. mc4100, Thermus thermophilus, Synthetic construct, Streptomyces sp., Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-01-28 Classification: Ribosome/antibiotic Ligands: MG, ZN, SF4, GDP |
 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Elongation Factor G In The Post-Translocational State (Without Fusitic Acid)
Organism: Escherichia coli str. k-12 substr. mc4100, Thermus thermophilus, Synthetic construct, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-01-28 Classification: RIBOSOME Ligands: MG, K, ZN, SF4, GDP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2014-11-26 Classification: OXIDOREDUCTASE Ligands: FOL, NAP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2014-11-26 Classification: OXIDOREDUCTASE Ligands: FOL, NAP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-11-26 Classification: OXIDOREDUCTASE Ligands: FOL, NAP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2014-10-15 Classification: OXIDOREDUCTASE Ligands: FOL, NAP, MN |
 |
Crystal Structure Of Rhomboid Intramembrane Protease Glpg F146I In Complex With Peptide Derived Inhibitor Ac-Fata-Cmk
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2014-09-24 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CL, BNG |
 |
Crystal Structure Of Rhomboid Intramembrane Protease Glpg In Complex With Peptide Derived Inhibitor Ac-Fata-Cmk
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2014-09-24 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CL, BNG |
 |
Crystal Structure Of Rhomboid Intramembrane Protease Glpg In Complex With Peptide Derived Inhibitor Ac-Iata-Cmk
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-09-24 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: BNG, CL |
 |
Structure Of The Curli Transport Lipoprotein Csgg In A Non-Lipidated, Pre-Pore Conformation
Organism: Escherichia coli str. k-12 substr. mc4100
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-09-24 Classification: TRANSPORT PROTEIN |
 |
Structure Of The Curli Transport Lipoprotein Csgg In Its Membrane- Bound Conformation
Organism: Escherichia coli str. k-12 substr. mc4100
Method: X-RAY DIFFRACTION Resolution:3.59 Å Release Date: 2014-09-24 Classification: TRANSPORT PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2014-07-23 Classification: ISOMERASE/ISOMERASE INHIBITOR Ligands: ZN |

